
Alteromonas confluentis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Alteromonas
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
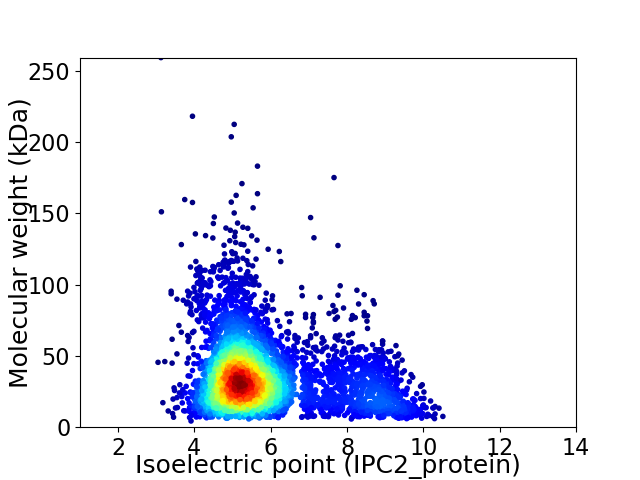
Virtual 2D-PAGE plot for 4086 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
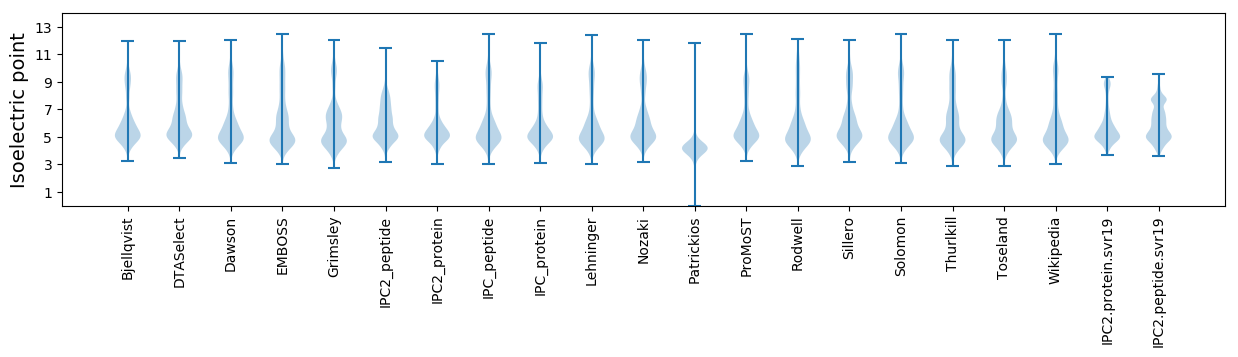
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E7ZF31|A0A1E7ZF31_9ALTE 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase OS=Alteromonas confluentis OX=1656094 GN=BFC18_04945 PE=3 SV=1
MM1 pKa = 7.69KK2 pKa = 10.5APIVLPLVLGITASVSASGSDD23 pKa = 3.59LLISQYY29 pKa = 11.15VEE31 pKa = 4.04GSSSNKK37 pKa = 9.88ALEE40 pKa = 4.7LYY42 pKa = 10.78NPTTEE47 pKa = 4.27SVSLSGYY54 pKa = 8.36TISVYY59 pKa = 11.31NNGGLSASSIVDD71 pKa = 3.53LSGSIAPGGTLVIAQSGAEE90 pKa = 4.13DD91 pKa = 3.77ALKK94 pKa = 10.86ALADD98 pKa = 3.82EE99 pKa = 4.87LTTQSLYY106 pKa = 11.27NGDD109 pKa = 3.88DD110 pKa = 4.28AIILTDD116 pKa = 3.73DD117 pKa = 4.28AGNILDD123 pKa = 5.06SIGQLGVDD131 pKa = 4.49PGSSWSEE138 pKa = 3.81GGLSTKK144 pKa = 10.57DD145 pKa = 2.83QTLIRR150 pKa = 11.84NSSVTTGDD158 pKa = 3.45TNPNDD163 pKa = 3.69AYY165 pKa = 10.87GLGEE169 pKa = 4.38WVGLPQNDD177 pKa = 3.62YY178 pKa = 10.45TGLGSHH184 pKa = 7.49DD185 pKa = 4.09YY186 pKa = 11.28DD187 pKa = 5.43GGGDD191 pKa = 3.68GDD193 pKa = 4.03GGGGEE198 pKa = 4.89DD199 pKa = 3.89EE200 pKa = 5.25DD201 pKa = 5.32LGGVCTNCPDD211 pKa = 3.39IDD213 pKa = 3.99KK214 pKa = 11.12VADD217 pKa = 3.64ASTFDD222 pKa = 3.67PAVYY226 pKa = 9.63YY227 pKa = 10.75AAVQSEE233 pKa = 4.13IDD235 pKa = 3.79AGSDD239 pKa = 3.13TAIIRR244 pKa = 11.84QILSEE249 pKa = 4.48TISGQHH255 pKa = 5.38VLTYY259 pKa = 10.62SEE261 pKa = 4.2VWTALTEE268 pKa = 4.18TDD270 pKa = 4.33EE271 pKa = 6.11DD272 pKa = 4.22PANPDD277 pKa = 3.95NILLIYY283 pKa = 9.96SGNSIPKK290 pKa = 9.33SEE292 pKa = 4.51NGSGSDD298 pKa = 4.58SSQPDD303 pKa = 3.24YY304 pKa = 11.05WNRR307 pKa = 11.84EE308 pKa = 3.91HH309 pKa = 6.94SWPKK313 pKa = 9.64SHH315 pKa = 6.39GFSSSSAEE323 pKa = 3.9AYY325 pKa = 9.68TDD327 pKa = 3.47IQHH330 pKa = 6.86LRR332 pKa = 11.84PSDD335 pKa = 3.13ITVNSSRR342 pKa = 11.84GNLDD346 pKa = 3.0FDD348 pKa = 4.67YY349 pKa = 11.32SDD351 pKa = 4.35APLPEE356 pKa = 5.25APANRR361 pKa = 11.84RR362 pKa = 11.84DD363 pKa = 3.56SDD365 pKa = 3.82SFEE368 pKa = 4.21PRR370 pKa = 11.84DD371 pKa = 3.67AVKK374 pKa = 10.95GDD376 pKa = 3.42IARR379 pKa = 11.84QMLYY383 pKa = 9.72MDD385 pKa = 4.14IRR387 pKa = 11.84YY388 pKa = 9.12EE389 pKa = 4.15GAGSDD394 pKa = 3.85VTPDD398 pKa = 3.2LALVNRR404 pKa = 11.84ITSSGEE410 pKa = 3.36PSLGKK415 pKa = 10.51LCTLLAWNDD424 pKa = 3.65ADD426 pKa = 3.64PVDD429 pKa = 3.92VVEE432 pKa = 4.84TNRR435 pKa = 11.84QSAIYY440 pKa = 9.11EE441 pKa = 4.04YY442 pKa = 10.54QGNRR446 pKa = 11.84NPFVDD451 pKa = 3.48HH452 pKa = 7.04PEE454 pKa = 4.07WVSLFYY460 pKa = 11.14SADD463 pKa = 3.27SCDD466 pKa = 3.53AVEE469 pKa = 5.1PEE471 pKa = 4.41PTPDD475 pKa = 4.09PEE477 pKa = 4.65PEE479 pKa = 4.41PEE481 pKa = 4.25PEE483 pKa = 4.03PTPVPGGFSPLTLTGVFDD501 pKa = 4.58GPLSGGTPKK510 pKa = 10.8GIEE513 pKa = 3.82LFVNQNIDD521 pKa = 3.59DD522 pKa = 4.71LSTCGVGSANNGGGTDD538 pKa = 3.7GEE540 pKa = 4.72EE541 pKa = 4.04FTFPAVSAVAGQFIYY556 pKa = 10.28IASEE560 pKa = 4.05SAGFEE565 pKa = 4.2AFFGFAPDD573 pKa = 3.75YY574 pKa = 10.48TSGAMSINGDD584 pKa = 3.5DD585 pKa = 4.63AIEE588 pKa = 4.22VFCDD592 pKa = 3.9GEE594 pKa = 4.61VVDD597 pKa = 6.54LYY599 pKa = 11.8GDD601 pKa = 3.64INTDD605 pKa = 3.17GTGEE609 pKa = 4.02AWDD612 pKa = 4.95HH613 pKa = 6.18LDD615 pKa = 2.82SWAYY619 pKa = 10.25RR620 pKa = 11.84IAGGPSATFDD630 pKa = 3.4VANWEE635 pKa = 4.31VAGTNVFDD643 pKa = 5.33GEE645 pKa = 4.55TSNATAAVPFPIGTYY660 pKa = 7.05VTPATEE666 pKa = 4.58LFFSEE671 pKa = 4.68YY672 pKa = 10.22IEE674 pKa = 4.62GSSNNKK680 pKa = 9.6ALEE683 pKa = 4.08IVNLTSAPVDD693 pKa = 3.77LSAYY697 pKa = 9.22DD698 pKa = 3.34VQMFINGRR706 pKa = 11.84DD707 pKa = 3.57YY708 pKa = 11.48AGQTFTLSGTLAPGDD723 pKa = 3.88VYY725 pKa = 11.71VLANSGSDD733 pKa = 3.48SAILAVADD741 pKa = 4.06LTSGSLTFNGDD752 pKa = 3.32DD753 pKa = 5.18AIVLRR758 pKa = 11.84NNGEE762 pKa = 4.03IVDD765 pKa = 3.81SFGRR769 pKa = 11.84IGEE772 pKa = 4.43DD773 pKa = 3.44PGSYY777 pKa = 8.58WQGDD781 pKa = 3.67DD782 pKa = 4.3VRR784 pKa = 11.84TQNRR788 pKa = 11.84TLVRR792 pKa = 11.84NAAITEE798 pKa = 4.29GDD800 pKa = 3.44TEE802 pKa = 5.05AYY804 pKa = 10.56DD805 pKa = 3.76EE806 pKa = 4.96FDD808 pKa = 5.28PSLEE812 pKa = 3.83WDD814 pKa = 4.19VYY816 pKa = 11.47DD817 pKa = 4.04QDD819 pKa = 3.79TFQFLGAYY827 pKa = 9.27GSSDD831 pKa = 2.95GGGGDD836 pKa = 3.93EE837 pKa = 5.4IGVCGDD843 pKa = 3.15EE844 pKa = 3.94ATFISEE850 pKa = 4.34VQGSGAEE857 pKa = 4.15SPLAGQSVVIEE868 pKa = 4.61AIVTHH873 pKa = 5.55VTPALEE879 pKa = 4.28GFWVQEE885 pKa = 3.9EE886 pKa = 4.34AADD889 pKa = 4.35SDD891 pKa = 4.5GDD893 pKa = 3.85DD894 pKa = 3.7TTSEE898 pKa = 4.52GVFIASTSLIEE909 pKa = 4.3VVTEE913 pKa = 3.88GDD915 pKa = 3.93VIRR918 pKa = 11.84VAGNVTEE925 pKa = 4.93DD926 pKa = 3.33YY927 pKa = 11.4GRR929 pKa = 11.84TLVEE933 pKa = 4.05ATSDD937 pKa = 3.57ALICGTGAVTPVVISLPMEE956 pKa = 4.28SADD959 pKa = 4.04SFEE962 pKa = 4.96KK963 pKa = 10.6YY964 pKa = 10.13EE965 pKa = 4.19GMVVTSSANWTISNIYY981 pKa = 9.03PYY983 pKa = 11.05SDD985 pKa = 3.72FGEE988 pKa = 4.33VWVSTEE994 pKa = 3.74RR995 pKa = 11.84LFNPTQIHH1003 pKa = 6.2LPGTEE1008 pKa = 3.77ASAALAASNALDD1020 pKa = 4.24VLIIDD1025 pKa = 5.04DD1026 pKa = 4.36NADD1029 pKa = 3.48GYY1031 pKa = 11.25ASEE1034 pKa = 4.08WMLPEE1039 pKa = 5.54GGFNPDD1045 pKa = 2.65NTIRR1049 pKa = 11.84SGDD1052 pKa = 4.36LITGVTGVMDD1062 pKa = 3.99YY1063 pKa = 10.99GYY1065 pKa = 9.72SAYY1068 pKa = 10.32RR1069 pKa = 11.84IRR1071 pKa = 11.84PTEE1074 pKa = 4.01VPSLVAEE1081 pKa = 4.78NYY1083 pKa = 9.73RR1084 pKa = 11.84EE1085 pKa = 3.83SSPYY1089 pKa = 10.03VEE1091 pKa = 4.44EE1092 pKa = 5.02GNLKK1096 pKa = 9.6VASFNVLNLFNGDD1109 pKa = 3.35GNGEE1113 pKa = 4.21GFPTARR1119 pKa = 11.84GADD1122 pKa = 3.4SLSEE1126 pKa = 4.16YY1127 pKa = 10.13EE1128 pKa = 4.12RR1129 pKa = 11.84QLAKK1133 pKa = 10.1IVNALVEE1140 pKa = 3.98IDD1142 pKa = 4.03ADD1144 pKa = 3.87VLGLMEE1150 pKa = 5.01IEE1152 pKa = 4.21NDD1154 pKa = 3.52GFGPLSAVAQLTEE1167 pKa = 3.9ALNDD1171 pKa = 3.49EE1172 pKa = 4.79FGEE1175 pKa = 4.42GTYY1178 pKa = 10.37AYY1180 pKa = 10.28VDD1182 pKa = 3.22AGGDD1186 pKa = 3.51QLGTDD1191 pKa = 5.06AITSGILYY1199 pKa = 9.62RR1200 pKa = 11.84PAVVTPVGSPAILLEE1215 pKa = 4.41ANSIVDD1221 pKa = 3.43EE1222 pKa = 4.51EE1223 pKa = 5.07GPLFVDD1229 pKa = 3.57YY1230 pKa = 11.24GNRR1233 pKa = 11.84PSLSQLFTHH1242 pKa = 6.2VEE1244 pKa = 3.98SGKK1247 pKa = 8.57TFAVDD1252 pKa = 3.5VNHH1255 pKa = 6.89LKK1257 pKa = 10.89SKK1259 pKa = 10.5GSSSSCQEE1267 pKa = 4.89DD1268 pKa = 3.61GDD1270 pKa = 5.17LEE1272 pKa = 4.06QGYY1275 pKa = 9.65CNRR1278 pKa = 11.84KK1279 pKa = 6.7RR1280 pKa = 11.84TRR1282 pKa = 11.84AAIAITAFLEE1292 pKa = 3.98QTYY1295 pKa = 10.98GDD1297 pKa = 3.84TATILLGDD1305 pKa = 3.64YY1306 pKa = 10.12NAYY1309 pKa = 10.42AKK1311 pKa = 10.09EE1312 pKa = 4.2DD1313 pKa = 4.73PIQVLASAGYY1323 pKa = 8.55ADD1325 pKa = 5.12SVAEE1329 pKa = 4.1MKK1331 pKa = 10.98GNASYY1336 pKa = 10.83SYY1338 pKa = 10.86SYY1340 pKa = 11.21DD1341 pKa = 3.28GLAGTLDD1348 pKa = 3.68YY1349 pKa = 11.09MLVNQAAMDD1358 pKa = 3.57MMVDD1362 pKa = 3.42VNEE1365 pKa = 3.74WHH1367 pKa = 7.04INSDD1371 pKa = 3.31EE1372 pKa = 4.24PEE1374 pKa = 4.19FYY1376 pKa = 10.39DD1377 pKa = 4.28YY1378 pKa = 11.73NEE1380 pKa = 4.26EE1381 pKa = 3.98YY1382 pKa = 10.74KK1383 pKa = 10.53PASFLNEE1390 pKa = 3.74LVYY1393 pKa = 10.4RR1394 pKa = 11.84ASDD1397 pKa = 3.64HH1398 pKa = 7.11DD1399 pKa = 4.37PVIGTYY1405 pKa = 10.51LLEE1408 pKa = 5.49AEE1410 pKa = 4.7EE1411 pKa = 4.58EE1412 pKa = 4.45PVTGDD1417 pKa = 2.66WDD1419 pKa = 3.72GDD1421 pKa = 3.76GDD1423 pKa = 4.02VDD1425 pKa = 5.45RR1426 pKa = 11.84DD1427 pKa = 3.8DD1428 pKa = 3.51VTALMRR1434 pKa = 11.84AIFARR1439 pKa = 11.84QEE1441 pKa = 3.33IDD1443 pKa = 3.5MSFDD1447 pKa = 3.72LNNDD1451 pKa = 2.62GRR1453 pKa = 11.84VNARR1457 pKa = 11.84DD1458 pKa = 3.17IGAMRR1463 pKa = 11.84RR1464 pKa = 11.84MCTRR1468 pKa = 11.84RR1469 pKa = 11.84GCATEE1474 pKa = 4.55DD1475 pKa = 3.25ATPSGRR1481 pKa = 11.84GRR1483 pKa = 11.84TLASAVTTKK1492 pKa = 10.79AAAISSLLNRR1502 pKa = 11.84NRR1504 pKa = 3.36
MM1 pKa = 7.69KK2 pKa = 10.5APIVLPLVLGITASVSASGSDD23 pKa = 3.59LLISQYY29 pKa = 11.15VEE31 pKa = 4.04GSSSNKK37 pKa = 9.88ALEE40 pKa = 4.7LYY42 pKa = 10.78NPTTEE47 pKa = 4.27SVSLSGYY54 pKa = 8.36TISVYY59 pKa = 11.31NNGGLSASSIVDD71 pKa = 3.53LSGSIAPGGTLVIAQSGAEE90 pKa = 4.13DD91 pKa = 3.77ALKK94 pKa = 10.86ALADD98 pKa = 3.82EE99 pKa = 4.87LTTQSLYY106 pKa = 11.27NGDD109 pKa = 3.88DD110 pKa = 4.28AIILTDD116 pKa = 3.73DD117 pKa = 4.28AGNILDD123 pKa = 5.06SIGQLGVDD131 pKa = 4.49PGSSWSEE138 pKa = 3.81GGLSTKK144 pKa = 10.57DD145 pKa = 2.83QTLIRR150 pKa = 11.84NSSVTTGDD158 pKa = 3.45TNPNDD163 pKa = 3.69AYY165 pKa = 10.87GLGEE169 pKa = 4.38WVGLPQNDD177 pKa = 3.62YY178 pKa = 10.45TGLGSHH184 pKa = 7.49DD185 pKa = 4.09YY186 pKa = 11.28DD187 pKa = 5.43GGGDD191 pKa = 3.68GDD193 pKa = 4.03GGGGEE198 pKa = 4.89DD199 pKa = 3.89EE200 pKa = 5.25DD201 pKa = 5.32LGGVCTNCPDD211 pKa = 3.39IDD213 pKa = 3.99KK214 pKa = 11.12VADD217 pKa = 3.64ASTFDD222 pKa = 3.67PAVYY226 pKa = 9.63YY227 pKa = 10.75AAVQSEE233 pKa = 4.13IDD235 pKa = 3.79AGSDD239 pKa = 3.13TAIIRR244 pKa = 11.84QILSEE249 pKa = 4.48TISGQHH255 pKa = 5.38VLTYY259 pKa = 10.62SEE261 pKa = 4.2VWTALTEE268 pKa = 4.18TDD270 pKa = 4.33EE271 pKa = 6.11DD272 pKa = 4.22PANPDD277 pKa = 3.95NILLIYY283 pKa = 9.96SGNSIPKK290 pKa = 9.33SEE292 pKa = 4.51NGSGSDD298 pKa = 4.58SSQPDD303 pKa = 3.24YY304 pKa = 11.05WNRR307 pKa = 11.84EE308 pKa = 3.91HH309 pKa = 6.94SWPKK313 pKa = 9.64SHH315 pKa = 6.39GFSSSSAEE323 pKa = 3.9AYY325 pKa = 9.68TDD327 pKa = 3.47IQHH330 pKa = 6.86LRR332 pKa = 11.84PSDD335 pKa = 3.13ITVNSSRR342 pKa = 11.84GNLDD346 pKa = 3.0FDD348 pKa = 4.67YY349 pKa = 11.32SDD351 pKa = 4.35APLPEE356 pKa = 5.25APANRR361 pKa = 11.84RR362 pKa = 11.84DD363 pKa = 3.56SDD365 pKa = 3.82SFEE368 pKa = 4.21PRR370 pKa = 11.84DD371 pKa = 3.67AVKK374 pKa = 10.95GDD376 pKa = 3.42IARR379 pKa = 11.84QMLYY383 pKa = 9.72MDD385 pKa = 4.14IRR387 pKa = 11.84YY388 pKa = 9.12EE389 pKa = 4.15GAGSDD394 pKa = 3.85VTPDD398 pKa = 3.2LALVNRR404 pKa = 11.84ITSSGEE410 pKa = 3.36PSLGKK415 pKa = 10.51LCTLLAWNDD424 pKa = 3.65ADD426 pKa = 3.64PVDD429 pKa = 3.92VVEE432 pKa = 4.84TNRR435 pKa = 11.84QSAIYY440 pKa = 9.11EE441 pKa = 4.04YY442 pKa = 10.54QGNRR446 pKa = 11.84NPFVDD451 pKa = 3.48HH452 pKa = 7.04PEE454 pKa = 4.07WVSLFYY460 pKa = 11.14SADD463 pKa = 3.27SCDD466 pKa = 3.53AVEE469 pKa = 5.1PEE471 pKa = 4.41PTPDD475 pKa = 4.09PEE477 pKa = 4.65PEE479 pKa = 4.41PEE481 pKa = 4.25PEE483 pKa = 4.03PTPVPGGFSPLTLTGVFDD501 pKa = 4.58GPLSGGTPKK510 pKa = 10.8GIEE513 pKa = 3.82LFVNQNIDD521 pKa = 3.59DD522 pKa = 4.71LSTCGVGSANNGGGTDD538 pKa = 3.7GEE540 pKa = 4.72EE541 pKa = 4.04FTFPAVSAVAGQFIYY556 pKa = 10.28IASEE560 pKa = 4.05SAGFEE565 pKa = 4.2AFFGFAPDD573 pKa = 3.75YY574 pKa = 10.48TSGAMSINGDD584 pKa = 3.5DD585 pKa = 4.63AIEE588 pKa = 4.22VFCDD592 pKa = 3.9GEE594 pKa = 4.61VVDD597 pKa = 6.54LYY599 pKa = 11.8GDD601 pKa = 3.64INTDD605 pKa = 3.17GTGEE609 pKa = 4.02AWDD612 pKa = 4.95HH613 pKa = 6.18LDD615 pKa = 2.82SWAYY619 pKa = 10.25RR620 pKa = 11.84IAGGPSATFDD630 pKa = 3.4VANWEE635 pKa = 4.31VAGTNVFDD643 pKa = 5.33GEE645 pKa = 4.55TSNATAAVPFPIGTYY660 pKa = 7.05VTPATEE666 pKa = 4.58LFFSEE671 pKa = 4.68YY672 pKa = 10.22IEE674 pKa = 4.62GSSNNKK680 pKa = 9.6ALEE683 pKa = 4.08IVNLTSAPVDD693 pKa = 3.77LSAYY697 pKa = 9.22DD698 pKa = 3.34VQMFINGRR706 pKa = 11.84DD707 pKa = 3.57YY708 pKa = 11.48AGQTFTLSGTLAPGDD723 pKa = 3.88VYY725 pKa = 11.71VLANSGSDD733 pKa = 3.48SAILAVADD741 pKa = 4.06LTSGSLTFNGDD752 pKa = 3.32DD753 pKa = 5.18AIVLRR758 pKa = 11.84NNGEE762 pKa = 4.03IVDD765 pKa = 3.81SFGRR769 pKa = 11.84IGEE772 pKa = 4.43DD773 pKa = 3.44PGSYY777 pKa = 8.58WQGDD781 pKa = 3.67DD782 pKa = 4.3VRR784 pKa = 11.84TQNRR788 pKa = 11.84TLVRR792 pKa = 11.84NAAITEE798 pKa = 4.29GDD800 pKa = 3.44TEE802 pKa = 5.05AYY804 pKa = 10.56DD805 pKa = 3.76EE806 pKa = 4.96FDD808 pKa = 5.28PSLEE812 pKa = 3.83WDD814 pKa = 4.19VYY816 pKa = 11.47DD817 pKa = 4.04QDD819 pKa = 3.79TFQFLGAYY827 pKa = 9.27GSSDD831 pKa = 2.95GGGGDD836 pKa = 3.93EE837 pKa = 5.4IGVCGDD843 pKa = 3.15EE844 pKa = 3.94ATFISEE850 pKa = 4.34VQGSGAEE857 pKa = 4.15SPLAGQSVVIEE868 pKa = 4.61AIVTHH873 pKa = 5.55VTPALEE879 pKa = 4.28GFWVQEE885 pKa = 3.9EE886 pKa = 4.34AADD889 pKa = 4.35SDD891 pKa = 4.5GDD893 pKa = 3.85DD894 pKa = 3.7TTSEE898 pKa = 4.52GVFIASTSLIEE909 pKa = 4.3VVTEE913 pKa = 3.88GDD915 pKa = 3.93VIRR918 pKa = 11.84VAGNVTEE925 pKa = 4.93DD926 pKa = 3.33YY927 pKa = 11.4GRR929 pKa = 11.84TLVEE933 pKa = 4.05ATSDD937 pKa = 3.57ALICGTGAVTPVVISLPMEE956 pKa = 4.28SADD959 pKa = 4.04SFEE962 pKa = 4.96KK963 pKa = 10.6YY964 pKa = 10.13EE965 pKa = 4.19GMVVTSSANWTISNIYY981 pKa = 9.03PYY983 pKa = 11.05SDD985 pKa = 3.72FGEE988 pKa = 4.33VWVSTEE994 pKa = 3.74RR995 pKa = 11.84LFNPTQIHH1003 pKa = 6.2LPGTEE1008 pKa = 3.77ASAALAASNALDD1020 pKa = 4.24VLIIDD1025 pKa = 5.04DD1026 pKa = 4.36NADD1029 pKa = 3.48GYY1031 pKa = 11.25ASEE1034 pKa = 4.08WMLPEE1039 pKa = 5.54GGFNPDD1045 pKa = 2.65NTIRR1049 pKa = 11.84SGDD1052 pKa = 4.36LITGVTGVMDD1062 pKa = 3.99YY1063 pKa = 10.99GYY1065 pKa = 9.72SAYY1068 pKa = 10.32RR1069 pKa = 11.84IRR1071 pKa = 11.84PTEE1074 pKa = 4.01VPSLVAEE1081 pKa = 4.78NYY1083 pKa = 9.73RR1084 pKa = 11.84EE1085 pKa = 3.83SSPYY1089 pKa = 10.03VEE1091 pKa = 4.44EE1092 pKa = 5.02GNLKK1096 pKa = 9.6VASFNVLNLFNGDD1109 pKa = 3.35GNGEE1113 pKa = 4.21GFPTARR1119 pKa = 11.84GADD1122 pKa = 3.4SLSEE1126 pKa = 4.16YY1127 pKa = 10.13EE1128 pKa = 4.12RR1129 pKa = 11.84QLAKK1133 pKa = 10.1IVNALVEE1140 pKa = 3.98IDD1142 pKa = 4.03ADD1144 pKa = 3.87VLGLMEE1150 pKa = 5.01IEE1152 pKa = 4.21NDD1154 pKa = 3.52GFGPLSAVAQLTEE1167 pKa = 3.9ALNDD1171 pKa = 3.49EE1172 pKa = 4.79FGEE1175 pKa = 4.42GTYY1178 pKa = 10.37AYY1180 pKa = 10.28VDD1182 pKa = 3.22AGGDD1186 pKa = 3.51QLGTDD1191 pKa = 5.06AITSGILYY1199 pKa = 9.62RR1200 pKa = 11.84PAVVTPVGSPAILLEE1215 pKa = 4.41ANSIVDD1221 pKa = 3.43EE1222 pKa = 4.51EE1223 pKa = 5.07GPLFVDD1229 pKa = 3.57YY1230 pKa = 11.24GNRR1233 pKa = 11.84PSLSQLFTHH1242 pKa = 6.2VEE1244 pKa = 3.98SGKK1247 pKa = 8.57TFAVDD1252 pKa = 3.5VNHH1255 pKa = 6.89LKK1257 pKa = 10.89SKK1259 pKa = 10.5GSSSSCQEE1267 pKa = 4.89DD1268 pKa = 3.61GDD1270 pKa = 5.17LEE1272 pKa = 4.06QGYY1275 pKa = 9.65CNRR1278 pKa = 11.84KK1279 pKa = 6.7RR1280 pKa = 11.84TRR1282 pKa = 11.84AAIAITAFLEE1292 pKa = 3.98QTYY1295 pKa = 10.98GDD1297 pKa = 3.84TATILLGDD1305 pKa = 3.64YY1306 pKa = 10.12NAYY1309 pKa = 10.42AKK1311 pKa = 10.09EE1312 pKa = 4.2DD1313 pKa = 4.73PIQVLASAGYY1323 pKa = 8.55ADD1325 pKa = 5.12SVAEE1329 pKa = 4.1MKK1331 pKa = 10.98GNASYY1336 pKa = 10.83SYY1338 pKa = 10.86SYY1340 pKa = 11.21DD1341 pKa = 3.28GLAGTLDD1348 pKa = 3.68YY1349 pKa = 11.09MLVNQAAMDD1358 pKa = 3.57MMVDD1362 pKa = 3.42VNEE1365 pKa = 3.74WHH1367 pKa = 7.04INSDD1371 pKa = 3.31EE1372 pKa = 4.24PEE1374 pKa = 4.19FYY1376 pKa = 10.39DD1377 pKa = 4.28YY1378 pKa = 11.73NEE1380 pKa = 4.26EE1381 pKa = 3.98YY1382 pKa = 10.74KK1383 pKa = 10.53PASFLNEE1390 pKa = 3.74LVYY1393 pKa = 10.4RR1394 pKa = 11.84ASDD1397 pKa = 3.64HH1398 pKa = 7.11DD1399 pKa = 4.37PVIGTYY1405 pKa = 10.51LLEE1408 pKa = 5.49AEE1410 pKa = 4.7EE1411 pKa = 4.58EE1412 pKa = 4.45PVTGDD1417 pKa = 2.66WDD1419 pKa = 3.72GDD1421 pKa = 3.76GDD1423 pKa = 4.02VDD1425 pKa = 5.45RR1426 pKa = 11.84DD1427 pKa = 3.8DD1428 pKa = 3.51VTALMRR1434 pKa = 11.84AIFARR1439 pKa = 11.84QEE1441 pKa = 3.33IDD1443 pKa = 3.5MSFDD1447 pKa = 3.72LNNDD1451 pKa = 2.62GRR1453 pKa = 11.84VNARR1457 pKa = 11.84DD1458 pKa = 3.17IGAMRR1463 pKa = 11.84RR1464 pKa = 11.84MCTRR1468 pKa = 11.84RR1469 pKa = 11.84GCATEE1474 pKa = 4.55DD1475 pKa = 3.25ATPSGRR1481 pKa = 11.84GRR1483 pKa = 11.84TLASAVTTKK1492 pKa = 10.79AAAISSLLNRR1502 pKa = 11.84NRR1504 pKa = 3.36
Molecular weight: 159.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E7ZDC0|A0A1E7ZDC0_9ALTE Cobalamin biosynthesis protein CobQ OS=Alteromonas confluentis OX=1656094 GN=BFC18_07175 PE=4 SV=1
MM1 pKa = 7.1EE2 pKa = 4.19TRR4 pKa = 11.84KK5 pKa = 10.16FSVLTRR11 pKa = 11.84GEE13 pKa = 4.02RR14 pKa = 11.84GVIAGVCSGLSEE26 pKa = 4.09YY27 pKa = 10.9FGFRR31 pKa = 11.84KK32 pKa = 9.89NCLRR36 pKa = 11.84IAFVVFSFFFALPVMIYY53 pKa = 9.1LTLWLILPKK62 pKa = 10.65YY63 pKa = 6.89PTSQAMARR71 pKa = 11.84QLRR74 pKa = 11.84RR75 pKa = 11.84QAGRR79 pKa = 11.84LSHH82 pKa = 7.36DD83 pKa = 3.46KK84 pKa = 11.1
MM1 pKa = 7.1EE2 pKa = 4.19TRR4 pKa = 11.84KK5 pKa = 10.16FSVLTRR11 pKa = 11.84GEE13 pKa = 4.02RR14 pKa = 11.84GVIAGVCSGLSEE26 pKa = 4.09YY27 pKa = 10.9FGFRR31 pKa = 11.84KK32 pKa = 9.89NCLRR36 pKa = 11.84IAFVVFSFFFALPVMIYY53 pKa = 9.1LTLWLILPKK62 pKa = 10.65YY63 pKa = 6.89PTSQAMARR71 pKa = 11.84QLRR74 pKa = 11.84RR75 pKa = 11.84QAGRR79 pKa = 11.84LSHH82 pKa = 7.36DD83 pKa = 3.46KK84 pKa = 11.1
Molecular weight: 9.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1417638 |
37 |
2500 |
347.0 |
38.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.28 ± 0.038 | 0.941 ± 0.013 |
5.98 ± 0.04 | 6.225 ± 0.034 |
4.222 ± 0.027 | 7.033 ± 0.037 |
2.145 ± 0.018 | 5.849 ± 0.028 |
4.748 ± 0.035 | 9.93 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.64 ± 0.017 | 4.201 ± 0.024 |
4.092 ± 0.024 | 4.202 ± 0.028 |
4.743 ± 0.025 | 6.63 ± 0.029 |
5.625 ± 0.026 | 7.176 ± 0.026 |
1.279 ± 0.015 | 3.059 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
