
Beihai sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
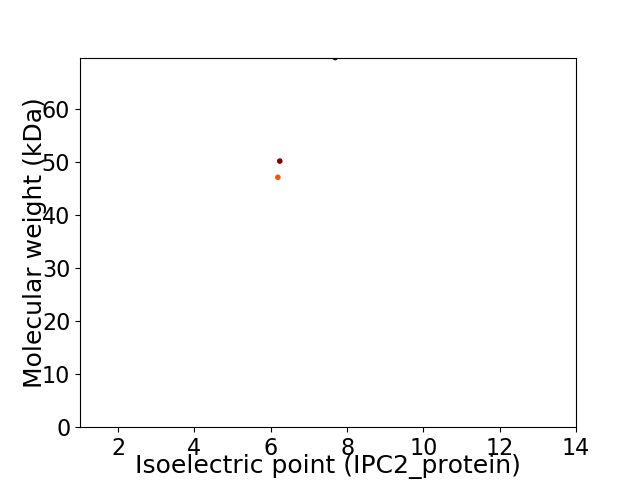
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
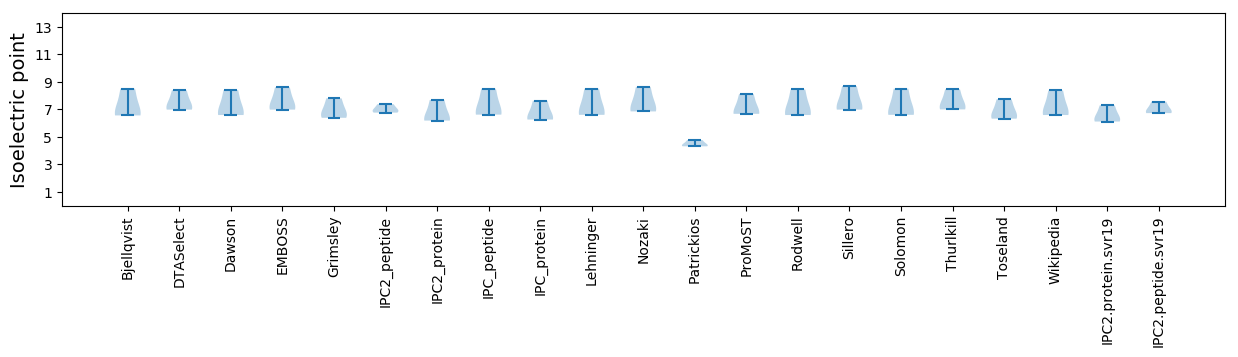
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KED6|A0A1L3KED6_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 1 OX=1922680 PE=4 SV=1
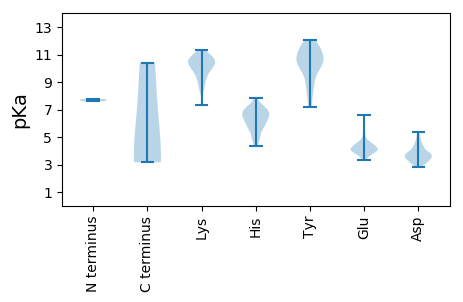
MM1 pKa = 7.68PSKK4 pKa = 9.8KK5 pKa = 8.65TRR7 pKa = 11.84EE8 pKa = 3.65IHH10 pKa = 5.57KK11 pKa = 9.9GEE13 pKa = 3.53RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.42RR24 pKa = 11.84NKK26 pKa = 9.63QLAHH30 pKa = 5.59GQKK33 pKa = 10.41GPQRR37 pKa = 11.84AQEE40 pKa = 4.16STPLRR45 pKa = 11.84DD46 pKa = 2.98VGSALGGIAGSLLGAPKK63 pKa = 10.05IGSLLGGALGSGVGKK78 pKa = 10.56LFGSGEE84 pKa = 3.92YY85 pKa = 10.36HH86 pKa = 6.71EE87 pKa = 5.5ALEE90 pKa = 4.59SEE92 pKa = 4.52LGAPVAEE99 pKa = 4.47PADD102 pKa = 3.75MPEE105 pKa = 4.32TNSLVEE111 pKa = 4.23PLSSNDD117 pKa = 3.91LVPLMHH123 pKa = 6.81QDD125 pKa = 3.11QEE127 pKa = 4.63GAVTITRR134 pKa = 11.84RR135 pKa = 11.84EE136 pKa = 4.16YY137 pKa = 10.54IRR139 pKa = 11.84DD140 pKa = 3.29ITILNSGSNFNFYY153 pKa = 10.14IGPGQQQFVWLSGIAKK169 pKa = 9.49SWQKK173 pKa = 10.42FAFTGLAMEE182 pKa = 4.31YY183 pKa = 10.57VPLSGTAVSSTSAALGQVAMCFSYY207 pKa = 11.15NVVEE211 pKa = 5.33STAAWPRR218 pKa = 11.84GVNAAILNMNGAVGCSPAAAATCYY242 pKa = 9.77MEE244 pKa = 5.19CDD246 pKa = 3.27PRR248 pKa = 11.84QAVNPFCFINNEE260 pKa = 3.95QPAVANWSEE269 pKa = 4.0SDD271 pKa = 3.68YY272 pKa = 11.24FPAEE276 pKa = 3.69FLLRR280 pKa = 11.84TSGAQEE286 pKa = 3.72LTTPFVCGALWCTYY300 pKa = 9.57EE301 pKa = 4.33VVLLEE306 pKa = 4.62PRR308 pKa = 11.84PVDD311 pKa = 3.57PSPALSFMNMPAFKK325 pKa = 10.84DD326 pKa = 4.0FVSVLNEE333 pKa = 3.47KK334 pKa = 10.31RR335 pKa = 11.84VLSNHH340 pKa = 6.43CGPYY344 pKa = 9.02TDD346 pKa = 3.57KK347 pKa = 11.23QVMLRR352 pKa = 11.84AAEE355 pKa = 4.06MRR357 pKa = 11.84RR358 pKa = 11.84LAAILRR364 pKa = 11.84SLPFADD370 pKa = 3.83ALEE373 pKa = 4.34KK374 pKa = 10.86ARR376 pKa = 11.84LQSWSIRR383 pKa = 11.84EE384 pKa = 4.09EE385 pKa = 4.35DD386 pKa = 4.14DD387 pKa = 3.78TATDD391 pKa = 3.31QQDD394 pKa = 3.54QVDD397 pKa = 5.4LIVSQLEE404 pKa = 4.09AKK406 pKa = 10.25YY407 pKa = 10.15KK408 pKa = 10.55ALSTEE413 pKa = 3.66MDD415 pKa = 3.2TWVQPVLTSTGSKK428 pKa = 8.98RR429 pKa = 11.84PKK431 pKa = 9.61YY432 pKa = 10.41
MM1 pKa = 7.68PSKK4 pKa = 9.8KK5 pKa = 8.65TRR7 pKa = 11.84EE8 pKa = 3.65IHH10 pKa = 5.57KK11 pKa = 9.9GEE13 pKa = 3.53RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.42RR24 pKa = 11.84NKK26 pKa = 9.63QLAHH30 pKa = 5.59GQKK33 pKa = 10.41GPQRR37 pKa = 11.84AQEE40 pKa = 4.16STPLRR45 pKa = 11.84DD46 pKa = 2.98VGSALGGIAGSLLGAPKK63 pKa = 10.05IGSLLGGALGSGVGKK78 pKa = 10.56LFGSGEE84 pKa = 3.92YY85 pKa = 10.36HH86 pKa = 6.71EE87 pKa = 5.5ALEE90 pKa = 4.59SEE92 pKa = 4.52LGAPVAEE99 pKa = 4.47PADD102 pKa = 3.75MPEE105 pKa = 4.32TNSLVEE111 pKa = 4.23PLSSNDD117 pKa = 3.91LVPLMHH123 pKa = 6.81QDD125 pKa = 3.11QEE127 pKa = 4.63GAVTITRR134 pKa = 11.84RR135 pKa = 11.84EE136 pKa = 4.16YY137 pKa = 10.54IRR139 pKa = 11.84DD140 pKa = 3.29ITILNSGSNFNFYY153 pKa = 10.14IGPGQQQFVWLSGIAKK169 pKa = 9.49SWQKK173 pKa = 10.42FAFTGLAMEE182 pKa = 4.31YY183 pKa = 10.57VPLSGTAVSSTSAALGQVAMCFSYY207 pKa = 11.15NVVEE211 pKa = 5.33STAAWPRR218 pKa = 11.84GVNAAILNMNGAVGCSPAAAATCYY242 pKa = 9.77MEE244 pKa = 5.19CDD246 pKa = 3.27PRR248 pKa = 11.84QAVNPFCFINNEE260 pKa = 3.95QPAVANWSEE269 pKa = 4.0SDD271 pKa = 3.68YY272 pKa = 11.24FPAEE276 pKa = 3.69FLLRR280 pKa = 11.84TSGAQEE286 pKa = 3.72LTTPFVCGALWCTYY300 pKa = 9.57EE301 pKa = 4.33VVLLEE306 pKa = 4.62PRR308 pKa = 11.84PVDD311 pKa = 3.57PSPALSFMNMPAFKK325 pKa = 10.84DD326 pKa = 4.0FVSVLNEE333 pKa = 3.47KK334 pKa = 10.31RR335 pKa = 11.84VLSNHH340 pKa = 6.43CGPYY344 pKa = 9.02TDD346 pKa = 3.57KK347 pKa = 11.23QVMLRR352 pKa = 11.84AAEE355 pKa = 4.06MRR357 pKa = 11.84RR358 pKa = 11.84LAAILRR364 pKa = 11.84SLPFADD370 pKa = 3.83ALEE373 pKa = 4.34KK374 pKa = 10.86ARR376 pKa = 11.84LQSWSIRR383 pKa = 11.84EE384 pKa = 4.09EE385 pKa = 4.35DD386 pKa = 4.14DD387 pKa = 3.78TATDD391 pKa = 3.31QQDD394 pKa = 3.54QVDD397 pKa = 5.4LIVSQLEE404 pKa = 4.09AKK406 pKa = 10.25YY407 pKa = 10.15KK408 pKa = 10.55ALSTEE413 pKa = 3.66MDD415 pKa = 3.2TWVQPVLTSTGSKK428 pKa = 8.98RR429 pKa = 11.84PKK431 pKa = 9.61YY432 pKa = 10.41
Molecular weight: 47.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KED8|A0A1L3KED8_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 1 OX=1922680 PE=4 SV=1
MM1 pKa = 7.59GPMEE5 pKa = 4.2MAVPRR10 pKa = 11.84LEE12 pKa = 4.67GDD14 pKa = 3.7TILIDD19 pKa = 3.79AGGEE23 pKa = 3.76IYY25 pKa = 10.29EE26 pKa = 4.44FSMTVTEE33 pKa = 5.67FSDD36 pKa = 4.45LVLKK40 pKa = 10.63QRR42 pKa = 11.84GRR44 pKa = 11.84MLFCEE49 pKa = 4.68RR50 pKa = 11.84AQEE53 pKa = 3.89LAKK56 pKa = 10.34PVIFDD61 pKa = 4.08PSPQYY66 pKa = 10.73RR67 pKa = 11.84RR68 pKa = 11.84VDD70 pKa = 3.7VKK72 pKa = 11.02EE73 pKa = 3.95STLPGSKK80 pKa = 9.92HH81 pKa = 6.63RR82 pKa = 11.84ILHH85 pKa = 5.72SLPNSCVQIHH95 pKa = 6.76AEE97 pKa = 4.08PGAHH101 pKa = 7.06RR102 pKa = 11.84LTGVHH107 pKa = 6.15GGGVRR112 pKa = 11.84VVYY115 pKa = 10.09GKK117 pKa = 8.44KK118 pKa = 8.39TYY120 pKa = 10.51LYY122 pKa = 8.35TAYY125 pKa = 10.31HH126 pKa = 5.98VIEE129 pKa = 5.0HH130 pKa = 7.0LLSQDD135 pKa = 3.51GVDD138 pKa = 4.5TICLRR143 pKa = 11.84ANGNDD148 pKa = 3.1VQMEE152 pKa = 3.93KK153 pKa = 10.9DD154 pKa = 3.35LRR156 pKa = 11.84FVAFSRR162 pKa = 11.84ALDD165 pKa = 3.64FVVIDD170 pKa = 3.64VPEE173 pKa = 4.24RR174 pKa = 11.84AWSKK178 pKa = 10.14LQVKK182 pKa = 9.13AAKK185 pKa = 9.46LAKK188 pKa = 9.42NWPSGAAVRR197 pKa = 11.84AIHH200 pKa = 7.1CDD202 pKa = 3.47GAGKK206 pKa = 10.61ASMAHH211 pKa = 7.06GYY213 pKa = 8.29LQSMEE218 pKa = 4.61KK219 pKa = 10.82GLLTHH224 pKa = 6.78TASTRR229 pKa = 11.84KK230 pKa = 9.78GSSGFPLMNLGGNLVYY246 pKa = 10.31GVHH249 pKa = 5.95VSGASTLNKK258 pKa = 9.71CSPAFFPFGGRR269 pKa = 11.84RR270 pKa = 11.84GASKK274 pKa = 10.66KK275 pKa = 10.54SADD278 pKa = 3.42GVLEE282 pKa = 3.99SSRR285 pKa = 11.84DD286 pKa = 3.75DD287 pKa = 3.9YY288 pKa = 11.77PDD290 pKa = 4.83DD291 pKa = 3.53EE292 pKa = 5.6LIRR295 pKa = 11.84EE296 pKa = 4.34FTYY299 pKa = 10.76EE300 pKa = 3.46EE301 pKa = 3.88WKK303 pKa = 10.8DD304 pKa = 3.05IEE306 pKa = 4.48RR307 pKa = 11.84YY308 pKa = 9.71HH309 pKa = 7.19IARR312 pKa = 11.84EE313 pKa = 4.02HH314 pKa = 6.45LGEE317 pKa = 5.16DD318 pKa = 3.8DD319 pKa = 3.72WDD321 pKa = 3.94DD322 pKa = 5.03FYY324 pKa = 11.83DD325 pKa = 3.9EE326 pKa = 4.64QLGGDD331 pKa = 4.08DD332 pKa = 4.69PYY334 pKa = 12.06ANDD337 pKa = 3.17FDD339 pKa = 4.28NRR341 pKa = 11.84EE342 pKa = 3.97EE343 pKa = 4.03VADD346 pKa = 4.18YY347 pKa = 11.54YY348 pKa = 11.56NALDD352 pKa = 3.77DD353 pKa = 4.94RR354 pKa = 11.84YY355 pKa = 11.05NEE357 pKa = 4.45AILRR361 pKa = 11.84FFGDD365 pKa = 3.05RR366 pKa = 11.84FVADD370 pKa = 4.39RR371 pKa = 11.84SGEE374 pKa = 4.01MTGRR378 pKa = 11.84EE379 pKa = 4.34SLNPAQYY386 pKa = 10.56AASVLANRR394 pKa = 11.84ARR396 pKa = 11.84HH397 pKa = 5.46VYY399 pKa = 10.5GFDD402 pKa = 3.25EE403 pKa = 6.64DD404 pKa = 4.37EE405 pKa = 4.61FEE407 pKa = 5.04SNGRR411 pKa = 11.84TIIVHH416 pKa = 6.41DD417 pKa = 4.38SKK419 pKa = 11.24ISGDD423 pKa = 3.26RR424 pKa = 11.84ALRR427 pKa = 11.84RR428 pKa = 11.84QEE430 pKa = 3.98NEE432 pKa = 3.38RR433 pKa = 11.84MADD436 pKa = 2.81IAAQRR441 pKa = 11.84KK442 pKa = 8.6ALLRR446 pKa = 11.84TRR448 pKa = 11.84DD449 pKa = 3.42QIASEE454 pKa = 3.92RR455 pKa = 11.84RR456 pKa = 11.84EE457 pKa = 4.15LEE459 pKa = 3.81RR460 pKa = 11.84QTRR463 pKa = 11.84SYY465 pKa = 11.26RR466 pKa = 11.84EE467 pKa = 3.35NLQRR471 pKa = 11.84KK472 pKa = 8.94RR473 pKa = 11.84EE474 pKa = 4.1ALKK477 pKa = 10.07EE478 pKa = 4.01AQIEE482 pKa = 4.06AQRR485 pKa = 11.84RR486 pKa = 11.84LKK488 pKa = 9.98EE489 pKa = 3.95LKK491 pKa = 9.17EE492 pKa = 4.05AKK494 pKa = 9.13TAFLKK499 pKa = 10.29EE500 pKa = 4.19HH501 pKa = 5.73EE502 pKa = 4.57AKK504 pKa = 10.59AAEE507 pKa = 4.42LDD509 pKa = 3.64RR510 pKa = 11.84AINAMSSSLPSEE522 pKa = 4.78SKK524 pKa = 10.99DD525 pKa = 3.15HH526 pKa = 4.82THH528 pKa = 6.09EE529 pKa = 4.17DD530 pKa = 3.3FRR532 pKa = 11.84RR533 pKa = 11.84RR534 pKa = 11.84LSPGQPEE541 pKa = 4.18PEE543 pKa = 4.18SPVKK547 pKa = 10.33GKK549 pKa = 10.52KK550 pKa = 7.43VTFQSQGQHH559 pKa = 4.99DD560 pKa = 4.51TKK562 pKa = 10.82NSKK565 pKa = 10.25PIEE568 pKa = 4.23TTQEE572 pKa = 4.07STGSKK577 pKa = 10.17APPSSSEE584 pKa = 3.95PKK586 pKa = 10.08RR587 pKa = 11.84PPLKK591 pKa = 9.81HH592 pKa = 6.46HH593 pKa = 6.69SPGSEE598 pKa = 3.68TGKK601 pKa = 10.15LPRR604 pKa = 11.84RR605 pKa = 11.84RR606 pKa = 11.84KK607 pKa = 9.46RR608 pKa = 11.84RR609 pKa = 11.84RR610 pKa = 11.84AASNRR615 pKa = 11.84KK616 pKa = 8.45KK617 pKa = 10.4QQ618 pKa = 3.25
MM1 pKa = 7.59GPMEE5 pKa = 4.2MAVPRR10 pKa = 11.84LEE12 pKa = 4.67GDD14 pKa = 3.7TILIDD19 pKa = 3.79AGGEE23 pKa = 3.76IYY25 pKa = 10.29EE26 pKa = 4.44FSMTVTEE33 pKa = 5.67FSDD36 pKa = 4.45LVLKK40 pKa = 10.63QRR42 pKa = 11.84GRR44 pKa = 11.84MLFCEE49 pKa = 4.68RR50 pKa = 11.84AQEE53 pKa = 3.89LAKK56 pKa = 10.34PVIFDD61 pKa = 4.08PSPQYY66 pKa = 10.73RR67 pKa = 11.84RR68 pKa = 11.84VDD70 pKa = 3.7VKK72 pKa = 11.02EE73 pKa = 3.95STLPGSKK80 pKa = 9.92HH81 pKa = 6.63RR82 pKa = 11.84ILHH85 pKa = 5.72SLPNSCVQIHH95 pKa = 6.76AEE97 pKa = 4.08PGAHH101 pKa = 7.06RR102 pKa = 11.84LTGVHH107 pKa = 6.15GGGVRR112 pKa = 11.84VVYY115 pKa = 10.09GKK117 pKa = 8.44KK118 pKa = 8.39TYY120 pKa = 10.51LYY122 pKa = 8.35TAYY125 pKa = 10.31HH126 pKa = 5.98VIEE129 pKa = 5.0HH130 pKa = 7.0LLSQDD135 pKa = 3.51GVDD138 pKa = 4.5TICLRR143 pKa = 11.84ANGNDD148 pKa = 3.1VQMEE152 pKa = 3.93KK153 pKa = 10.9DD154 pKa = 3.35LRR156 pKa = 11.84FVAFSRR162 pKa = 11.84ALDD165 pKa = 3.64FVVIDD170 pKa = 3.64VPEE173 pKa = 4.24RR174 pKa = 11.84AWSKK178 pKa = 10.14LQVKK182 pKa = 9.13AAKK185 pKa = 9.46LAKK188 pKa = 9.42NWPSGAAVRR197 pKa = 11.84AIHH200 pKa = 7.1CDD202 pKa = 3.47GAGKK206 pKa = 10.61ASMAHH211 pKa = 7.06GYY213 pKa = 8.29LQSMEE218 pKa = 4.61KK219 pKa = 10.82GLLTHH224 pKa = 6.78TASTRR229 pKa = 11.84KK230 pKa = 9.78GSSGFPLMNLGGNLVYY246 pKa = 10.31GVHH249 pKa = 5.95VSGASTLNKK258 pKa = 9.71CSPAFFPFGGRR269 pKa = 11.84RR270 pKa = 11.84GASKK274 pKa = 10.66KK275 pKa = 10.54SADD278 pKa = 3.42GVLEE282 pKa = 3.99SSRR285 pKa = 11.84DD286 pKa = 3.75DD287 pKa = 3.9YY288 pKa = 11.77PDD290 pKa = 4.83DD291 pKa = 3.53EE292 pKa = 5.6LIRR295 pKa = 11.84EE296 pKa = 4.34FTYY299 pKa = 10.76EE300 pKa = 3.46EE301 pKa = 3.88WKK303 pKa = 10.8DD304 pKa = 3.05IEE306 pKa = 4.48RR307 pKa = 11.84YY308 pKa = 9.71HH309 pKa = 7.19IARR312 pKa = 11.84EE313 pKa = 4.02HH314 pKa = 6.45LGEE317 pKa = 5.16DD318 pKa = 3.8DD319 pKa = 3.72WDD321 pKa = 3.94DD322 pKa = 5.03FYY324 pKa = 11.83DD325 pKa = 3.9EE326 pKa = 4.64QLGGDD331 pKa = 4.08DD332 pKa = 4.69PYY334 pKa = 12.06ANDD337 pKa = 3.17FDD339 pKa = 4.28NRR341 pKa = 11.84EE342 pKa = 3.97EE343 pKa = 4.03VADD346 pKa = 4.18YY347 pKa = 11.54YY348 pKa = 11.56NALDD352 pKa = 3.77DD353 pKa = 4.94RR354 pKa = 11.84YY355 pKa = 11.05NEE357 pKa = 4.45AILRR361 pKa = 11.84FFGDD365 pKa = 3.05RR366 pKa = 11.84FVADD370 pKa = 4.39RR371 pKa = 11.84SGEE374 pKa = 4.01MTGRR378 pKa = 11.84EE379 pKa = 4.34SLNPAQYY386 pKa = 10.56AASVLANRR394 pKa = 11.84ARR396 pKa = 11.84HH397 pKa = 5.46VYY399 pKa = 10.5GFDD402 pKa = 3.25EE403 pKa = 6.64DD404 pKa = 4.37EE405 pKa = 4.61FEE407 pKa = 5.04SNGRR411 pKa = 11.84TIIVHH416 pKa = 6.41DD417 pKa = 4.38SKK419 pKa = 11.24ISGDD423 pKa = 3.26RR424 pKa = 11.84ALRR427 pKa = 11.84RR428 pKa = 11.84QEE430 pKa = 3.98NEE432 pKa = 3.38RR433 pKa = 11.84MADD436 pKa = 2.81IAAQRR441 pKa = 11.84KK442 pKa = 8.6ALLRR446 pKa = 11.84TRR448 pKa = 11.84DD449 pKa = 3.42QIASEE454 pKa = 3.92RR455 pKa = 11.84RR456 pKa = 11.84EE457 pKa = 4.15LEE459 pKa = 3.81RR460 pKa = 11.84QTRR463 pKa = 11.84SYY465 pKa = 11.26RR466 pKa = 11.84EE467 pKa = 3.35NLQRR471 pKa = 11.84KK472 pKa = 8.94RR473 pKa = 11.84EE474 pKa = 4.1ALKK477 pKa = 10.07EE478 pKa = 4.01AQIEE482 pKa = 4.06AQRR485 pKa = 11.84RR486 pKa = 11.84LKK488 pKa = 9.98EE489 pKa = 3.95LKK491 pKa = 9.17EE492 pKa = 4.05AKK494 pKa = 9.13TAFLKK499 pKa = 10.29EE500 pKa = 4.19HH501 pKa = 5.73EE502 pKa = 4.57AKK504 pKa = 10.59AAEE507 pKa = 4.42LDD509 pKa = 3.64RR510 pKa = 11.84AINAMSSSLPSEE522 pKa = 4.78SKK524 pKa = 10.99DD525 pKa = 3.15HH526 pKa = 4.82THH528 pKa = 6.09EE529 pKa = 4.17DD530 pKa = 3.3FRR532 pKa = 11.84RR533 pKa = 11.84RR534 pKa = 11.84LSPGQPEE541 pKa = 4.18PEE543 pKa = 4.18SPVKK547 pKa = 10.33GKK549 pKa = 10.52KK550 pKa = 7.43VTFQSQGQHH559 pKa = 4.99DD560 pKa = 4.51TKK562 pKa = 10.82NSKK565 pKa = 10.25PIEE568 pKa = 4.23TTQEE572 pKa = 4.07STGSKK577 pKa = 10.17APPSSSEE584 pKa = 3.95PKK586 pKa = 10.08RR587 pKa = 11.84PPLKK591 pKa = 9.81HH592 pKa = 6.46HH593 pKa = 6.69SPGSEE598 pKa = 3.68TGKK601 pKa = 10.15LPRR604 pKa = 11.84RR605 pKa = 11.84RR606 pKa = 11.84KK607 pKa = 9.46RR608 pKa = 11.84RR609 pKa = 11.84RR610 pKa = 11.84AASNRR615 pKa = 11.84KK616 pKa = 8.45KK617 pKa = 10.4QQ618 pKa = 3.25
Molecular weight: 69.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1496 |
432 |
618 |
498.7 |
55.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.024 ± 0.699 | 1.537 ± 0.386 |
5.882 ± 0.641 | 7.286 ± 0.567 |
3.275 ± 0.307 | 6.885 ± 0.314 |
2.607 ± 0.493 | 3.743 ± 0.281 |
5.214 ± 0.626 | 8.556 ± 0.476 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.201 | 3.275 ± 0.216 |
5.414 ± 0.367 | 4.011 ± 0.277 |
8.021 ± 0.747 | 7.888 ± 0.148 |
4.479 ± 0.234 | 6.417 ± 0.842 |
1.404 ± 0.418 | 2.741 ± 0.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
