
Marinobacter phage PS6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
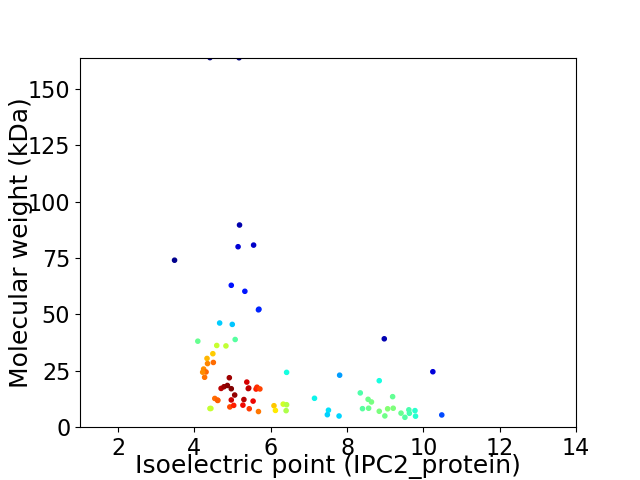
Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2D1GMX0|A0A2D1GMX0_9CAUD Uncharacterized protein OS=Marinobacter phage PS6 OX=2041342 PE=4 SV=1
MM1 pKa = 7.75ASILGKK7 pKa = 10.34LAIQEE12 pKa = 4.24GDD14 pKa = 3.63SYY16 pKa = 11.13TAAAGSNRR24 pKa = 11.84AVVVAGTGEE33 pKa = 4.22SAFSSRR39 pKa = 11.84EE40 pKa = 3.51IVGITFGGVAMTRR53 pKa = 11.84AAYY56 pKa = 9.13TGDD59 pKa = 3.4TEE61 pKa = 4.34TAKK64 pKa = 11.02GGVYY68 pKa = 10.39YY69 pKa = 9.99ILEE72 pKa = 4.19SQIPAGSSVIAVDD85 pKa = 3.06WDD87 pKa = 4.15GQVHH91 pKa = 6.65DD92 pKa = 5.02AFTGAVYY99 pKa = 9.74TLGGIDD105 pKa = 3.85QATPVNNFNSAVGVSLFDD123 pKa = 4.62LDD125 pKa = 4.19IMFNGVDD132 pKa = 3.6GGVLIVGAGSNSPADD147 pKa = 3.83TLNQSSTISATTTTVTVDD165 pKa = 3.36HH166 pKa = 6.7AQTTAGHH173 pKa = 6.31EE174 pKa = 4.37GSAISGVLSAAGSEE188 pKa = 4.43TLGLRR193 pKa = 11.84YY194 pKa = 9.81SSEE197 pKa = 4.03TDD199 pKa = 3.25PVGALAVFNPATSGPSVSAADD220 pKa = 5.14DD221 pKa = 3.5ITSEE225 pKa = 4.05GDD227 pKa = 3.45TVEE230 pKa = 4.67FTLADD235 pKa = 3.46NSAAPVSATLNGTDD249 pKa = 3.31VGTLTLVSGSTYY261 pKa = 10.69SYY263 pKa = 8.06TAPLIADD270 pKa = 4.93DD271 pKa = 4.29DD272 pKa = 4.27TADD275 pKa = 3.85LVVSVDD281 pKa = 3.16STTASTVISYY291 pKa = 10.96ANSYY295 pKa = 9.75PYY297 pKa = 10.77EE298 pKa = 4.28LVTHH302 pKa = 6.53GEE304 pKa = 3.87PDD306 pKa = 3.3ANSAFFDD313 pKa = 3.72TAFATDD319 pKa = 5.15GPVEE323 pKa = 4.21WGVVTDD329 pKa = 4.29FDD331 pKa = 4.22SGVVVVDD338 pKa = 3.89WAAMDD343 pKa = 3.88AAEE346 pKa = 6.02DD347 pKa = 3.98EE348 pKa = 4.82LNDD351 pKa = 3.49IALHH355 pKa = 4.6STEE358 pKa = 4.18VAAGDD363 pKa = 3.56STATLKK369 pKa = 11.1YY370 pKa = 9.44FVPEE374 pKa = 4.12SGATGTFQATATVDD388 pKa = 3.81GTDD391 pKa = 3.3EE392 pKa = 4.01TAPTISSASVPTAGNNIAVQMSEE415 pKa = 3.9SMQVGAGGSGGWTISLGDD433 pKa = 3.65ATVTSAAVDD442 pKa = 3.99GSDD445 pKa = 3.37DD446 pKa = 3.65TTVNLTPSRR455 pKa = 11.84TLTDD459 pKa = 3.41EE460 pKa = 4.01DD461 pKa = 4.96TFTIGYY467 pKa = 5.39TQPGNGFQDD476 pKa = 3.77QADD479 pKa = 4.34TPNDD483 pKa = 3.51LATLSGQVVTNNSTQQPPDD502 pKa = 3.08VTGPVIVSVSVPTTGTYY519 pKa = 10.89APGDD523 pKa = 3.61ALNFTVNFDD532 pKa = 3.66EE533 pKa = 4.25NTFRR537 pKa = 11.84TGVPALNLDD546 pKa = 3.67VGGSARR552 pKa = 11.84QANYY556 pKa = 10.53VSGTDD561 pKa = 3.15TTALVFRR568 pKa = 11.84YY569 pKa = 7.21TVQEE573 pKa = 3.98GDD575 pKa = 3.72EE576 pKa = 4.31DD577 pKa = 4.29TDD579 pKa = 4.89GITVSSLTLDD589 pKa = 3.43GGTLKK594 pKa = 10.99DD595 pKa = 3.59SAGNDD600 pKa = 3.04ATLTLNGVGDD610 pKa = 3.78TSGILIDD617 pKa = 4.5GVSPTISIGAINTTDD632 pKa = 3.44TTPTASGSAGDD643 pKa = 4.27SDD645 pKa = 5.75SLTLDD650 pKa = 3.42LDD652 pKa = 4.39GVDD655 pKa = 3.8VTHH658 pKa = 6.82TSSYY662 pKa = 11.09NITPSGGSWSQQFPEE677 pKa = 4.57LAIGTYY683 pKa = 9.68TMTLTGVDD691 pKa = 3.25TAGNEE696 pKa = 4.36TVEE699 pKa = 4.31TATLQIVEE707 pKa = 4.77PSTSPSSKK715 pKa = 10.5LLRR718 pKa = 11.84QLLRR722 pKa = 11.84GSLRR726 pKa = 11.84SSLKK730 pKa = 10.73LSS732 pKa = 3.51
MM1 pKa = 7.75ASILGKK7 pKa = 10.34LAIQEE12 pKa = 4.24GDD14 pKa = 3.63SYY16 pKa = 11.13TAAAGSNRR24 pKa = 11.84AVVVAGTGEE33 pKa = 4.22SAFSSRR39 pKa = 11.84EE40 pKa = 3.51IVGITFGGVAMTRR53 pKa = 11.84AAYY56 pKa = 9.13TGDD59 pKa = 3.4TEE61 pKa = 4.34TAKK64 pKa = 11.02GGVYY68 pKa = 10.39YY69 pKa = 9.99ILEE72 pKa = 4.19SQIPAGSSVIAVDD85 pKa = 3.06WDD87 pKa = 4.15GQVHH91 pKa = 6.65DD92 pKa = 5.02AFTGAVYY99 pKa = 9.74TLGGIDD105 pKa = 3.85QATPVNNFNSAVGVSLFDD123 pKa = 4.62LDD125 pKa = 4.19IMFNGVDD132 pKa = 3.6GGVLIVGAGSNSPADD147 pKa = 3.83TLNQSSTISATTTTVTVDD165 pKa = 3.36HH166 pKa = 6.7AQTTAGHH173 pKa = 6.31EE174 pKa = 4.37GSAISGVLSAAGSEE188 pKa = 4.43TLGLRR193 pKa = 11.84YY194 pKa = 9.81SSEE197 pKa = 4.03TDD199 pKa = 3.25PVGALAVFNPATSGPSVSAADD220 pKa = 5.14DD221 pKa = 3.5ITSEE225 pKa = 4.05GDD227 pKa = 3.45TVEE230 pKa = 4.67FTLADD235 pKa = 3.46NSAAPVSATLNGTDD249 pKa = 3.31VGTLTLVSGSTYY261 pKa = 10.69SYY263 pKa = 8.06TAPLIADD270 pKa = 4.93DD271 pKa = 4.29DD272 pKa = 4.27TADD275 pKa = 3.85LVVSVDD281 pKa = 3.16STTASTVISYY291 pKa = 10.96ANSYY295 pKa = 9.75PYY297 pKa = 10.77EE298 pKa = 4.28LVTHH302 pKa = 6.53GEE304 pKa = 3.87PDD306 pKa = 3.3ANSAFFDD313 pKa = 3.72TAFATDD319 pKa = 5.15GPVEE323 pKa = 4.21WGVVTDD329 pKa = 4.29FDD331 pKa = 4.22SGVVVVDD338 pKa = 3.89WAAMDD343 pKa = 3.88AAEE346 pKa = 6.02DD347 pKa = 3.98EE348 pKa = 4.82LNDD351 pKa = 3.49IALHH355 pKa = 4.6STEE358 pKa = 4.18VAAGDD363 pKa = 3.56STATLKK369 pKa = 11.1YY370 pKa = 9.44FVPEE374 pKa = 4.12SGATGTFQATATVDD388 pKa = 3.81GTDD391 pKa = 3.3EE392 pKa = 4.01TAPTISSASVPTAGNNIAVQMSEE415 pKa = 3.9SMQVGAGGSGGWTISLGDD433 pKa = 3.65ATVTSAAVDD442 pKa = 3.99GSDD445 pKa = 3.37DD446 pKa = 3.65TTVNLTPSRR455 pKa = 11.84TLTDD459 pKa = 3.41EE460 pKa = 4.01DD461 pKa = 4.96TFTIGYY467 pKa = 5.39TQPGNGFQDD476 pKa = 3.77QADD479 pKa = 4.34TPNDD483 pKa = 3.51LATLSGQVVTNNSTQQPPDD502 pKa = 3.08VTGPVIVSVSVPTTGTYY519 pKa = 10.89APGDD523 pKa = 3.61ALNFTVNFDD532 pKa = 3.66EE533 pKa = 4.25NTFRR537 pKa = 11.84TGVPALNLDD546 pKa = 3.67VGGSARR552 pKa = 11.84QANYY556 pKa = 10.53VSGTDD561 pKa = 3.15TTALVFRR568 pKa = 11.84YY569 pKa = 7.21TVQEE573 pKa = 3.98GDD575 pKa = 3.72EE576 pKa = 4.31DD577 pKa = 4.29TDD579 pKa = 4.89GITVSSLTLDD589 pKa = 3.43GGTLKK594 pKa = 10.99DD595 pKa = 3.59SAGNDD600 pKa = 3.04ATLTLNGVGDD610 pKa = 3.78TSGILIDD617 pKa = 4.5GVSPTISIGAINTTDD632 pKa = 3.44TTPTASGSAGDD643 pKa = 4.27SDD645 pKa = 5.75SLTLDD650 pKa = 3.42LDD652 pKa = 4.39GVDD655 pKa = 3.8VTHH658 pKa = 6.82TSSYY662 pKa = 11.09NITPSGGSWSQQFPEE677 pKa = 4.57LAIGTYY683 pKa = 9.68TMTLTGVDD691 pKa = 3.25TAGNEE696 pKa = 4.36TVEE699 pKa = 4.31TATLQIVEE707 pKa = 4.77PSTSPSSKK715 pKa = 10.5LLRR718 pKa = 11.84QLLRR722 pKa = 11.84GSLRR726 pKa = 11.84SSLKK730 pKa = 10.73LSS732 pKa = 3.51
Molecular weight: 74.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1GMP0|A0A2D1GMP0_9CAUD Uncharacterized protein OS=Marinobacter phage PS6 OX=2041342 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84HH3 pKa = 5.47MGYY6 pKa = 9.98EE7 pKa = 3.83PRR9 pKa = 11.84EE10 pKa = 3.59IWEE13 pKa = 4.67DD14 pKa = 3.65LNEE17 pKa = 4.07QPHH20 pKa = 5.97IVRR23 pKa = 11.84KK24 pKa = 10.04IISDD28 pKa = 3.37HH29 pKa = 5.24VNRR32 pKa = 11.84YY33 pKa = 8.65SPEE36 pKa = 3.67RR37 pKa = 11.84VEE39 pKa = 6.15AIRR42 pKa = 11.84QTRR45 pKa = 11.84KK46 pKa = 9.32RR47 pKa = 11.84QRR49 pKa = 11.84QNKK52 pKa = 6.7ATRR55 pKa = 11.84KK56 pKa = 9.66RR57 pKa = 11.84LAEE60 pKa = 3.95QQQ62 pKa = 3.2
MM1 pKa = 7.55RR2 pKa = 11.84HH3 pKa = 5.47MGYY6 pKa = 9.98EE7 pKa = 3.83PRR9 pKa = 11.84EE10 pKa = 3.59IWEE13 pKa = 4.67DD14 pKa = 3.65LNEE17 pKa = 4.07QPHH20 pKa = 5.97IVRR23 pKa = 11.84KK24 pKa = 10.04IISDD28 pKa = 3.37HH29 pKa = 5.24VNRR32 pKa = 11.84YY33 pKa = 8.65SPEE36 pKa = 3.67RR37 pKa = 11.84VEE39 pKa = 6.15AIRR42 pKa = 11.84QTRR45 pKa = 11.84KK46 pKa = 9.32RR47 pKa = 11.84QRR49 pKa = 11.84QNKK52 pKa = 6.7ATRR55 pKa = 11.84KK56 pKa = 9.66RR57 pKa = 11.84LAEE60 pKa = 3.95QQQ62 pKa = 3.2
Molecular weight: 7.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18163 |
41 |
1500 |
227.0 |
25.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.526 ± 0.474 | 0.864 ± 0.117 |
7.086 ± 0.276 | 7.185 ± 0.411 |
4.019 ± 0.185 | 8.016 ± 0.344 |
1.767 ± 0.163 | 5.049 ± 0.171 |
5.296 ± 0.367 | 7.741 ± 0.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.209 | 4.454 ± 0.173 |
4.184 ± 0.264 | 4.008 ± 0.33 |
5.963 ± 0.386 | 6.172 ± 0.293 |
6.524 ± 0.432 | 6.794 ± 0.301 |
1.63 ± 0.156 | 3.254 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
