
Sediminihabitans luteus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Sediminihabitans
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
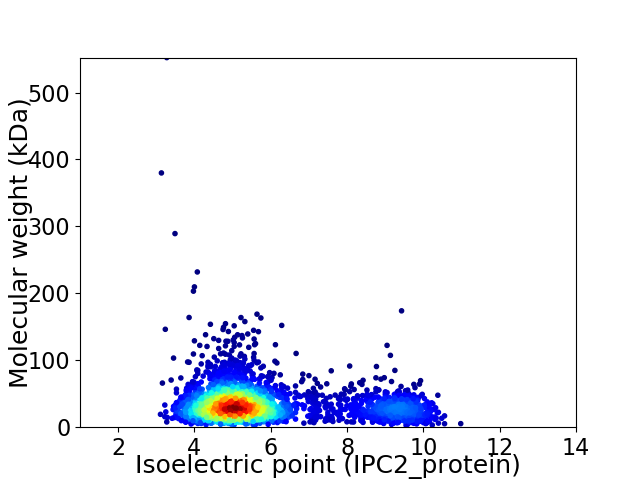
Virtual 2D-PAGE plot for 2995 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9D055|A0A2M9D055_9CELL Uncharacterized protein OS=Sediminihabitans luteus OX=1138585 GN=CLV28_0806 PE=4 SV=1
MM1 pKa = 6.85PTHH4 pKa = 5.73YY5 pKa = 10.14RR6 pKa = 11.84YY7 pKa = 9.96GVSMRR12 pKa = 11.84TSRR15 pKa = 11.84PATVAVVVAALVTALLAGCDD35 pKa = 3.79APGKK39 pKa = 9.33EE40 pKa = 4.34QTPTLDD46 pKa = 3.55PSASSTQGADD56 pKa = 3.53PLAGAPAGYY65 pKa = 9.9EE66 pKa = 3.92SYY68 pKa = 11.01YY69 pKa = 10.88GQTLEE74 pKa = 4.39WEE76 pKa = 4.59SCGGGYY82 pKa = 10.3EE83 pKa = 4.38CTDD86 pKa = 3.41ADD88 pKa = 5.92APISWQDD95 pKa = 3.2PDD97 pKa = 4.26LGSIQLALKK106 pKa = 10.02RR107 pKa = 11.84LPASGDD113 pKa = 3.48RR114 pKa = 11.84QGSLLINPGGPGSSGVEE131 pKa = 4.14FVTSAASIIGDD142 pKa = 3.89RR143 pKa = 11.84VKK145 pKa = 10.79GAYY148 pKa = 9.88DD149 pKa = 3.44LVGFDD154 pKa = 3.75PRR156 pKa = 11.84GVGGSAPVKK165 pKa = 10.7CLDD168 pKa = 5.2DD169 pKa = 4.36ADD171 pKa = 4.6KK172 pKa = 11.17DD173 pKa = 3.88ASLSASYY180 pKa = 9.94PHH182 pKa = 5.69TTEE185 pKa = 5.62GIAAMADD192 pKa = 3.75DD193 pKa = 4.69LEE195 pKa = 5.26AWAQACDD202 pKa = 3.83EE203 pKa = 4.3NTGEE207 pKa = 4.37QLATVDD213 pKa = 3.81TQSAARR219 pKa = 11.84DD220 pKa = 3.49MDD222 pKa = 3.72MLRR225 pKa = 11.84AVLGDD230 pKa = 3.73EE231 pKa = 3.95QLHH234 pKa = 5.52YY235 pKa = 11.17LGFSYY240 pKa = 8.67GTQLGATYY248 pKa = 10.71AGLFPQTVGRR258 pKa = 11.84MVLDD262 pKa = 3.92GAIDD266 pKa = 3.58TTLDD270 pKa = 3.31ADD272 pKa = 4.53EE273 pKa = 5.19ISAEE277 pKa = 3.97QAVGFEE283 pKa = 4.25NALRR287 pKa = 11.84AYY289 pKa = 9.2VTDD292 pKa = 3.9CLGGRR297 pKa = 11.84ACPLSGSVDD306 pKa = 2.83GGMQQIRR313 pKa = 11.84DD314 pKa = 3.76LLDD317 pKa = 3.37RR318 pKa = 11.84TLEE321 pKa = 4.47DD322 pKa = 4.48PLPTDD327 pKa = 3.31SDD329 pKa = 3.86RR330 pKa = 11.84DD331 pKa = 3.7LTQSLTFYY339 pKa = 11.13GIALPLYY346 pKa = 10.64SKK348 pKa = 11.23ANWSLLTNALSDD360 pKa = 3.76ALEE363 pKa = 5.1DD364 pKa = 4.3GDD366 pKa = 5.63GSTLLYY372 pKa = 10.81LADD375 pKa = 5.28FYY377 pKa = 11.77NDD379 pKa = 3.72RR380 pKa = 11.84QPDD383 pKa = 3.58GSFSSNSAEE392 pKa = 3.77AFRR395 pKa = 11.84AINCLDD401 pKa = 3.5DD402 pKa = 5.18RR403 pKa = 11.84GNPDD407 pKa = 3.89PDD409 pKa = 3.9FMAEE413 pKa = 3.72QAEE416 pKa = 4.71EE417 pKa = 4.15IEE419 pKa = 4.39AAAPTMGEE427 pKa = 3.92FFTYY431 pKa = 10.4SGLTCADD438 pKa = 3.23WPVPEE443 pKa = 5.07VEE445 pKa = 4.37QQFDD449 pKa = 3.43LHH451 pKa = 7.65ASGAAPIVVVGTTNDD466 pKa = 3.55PATPYY471 pKa = 10.5VWAQGLAKK479 pKa = 9.67TLDD482 pKa = 3.54SATLVTFEE490 pKa = 4.97GEE492 pKa = 3.74GHH494 pKa = 5.44TAYY497 pKa = 10.31GSSNDD502 pKa = 3.9CVGDD506 pKa = 3.61AVDD509 pKa = 5.52DD510 pKa = 4.03YY511 pKa = 11.55LVDD514 pKa = 3.6GTVPKK519 pKa = 10.82DD520 pKa = 3.3GLTCC524 pKa = 4.55
MM1 pKa = 6.85PTHH4 pKa = 5.73YY5 pKa = 10.14RR6 pKa = 11.84YY7 pKa = 9.96GVSMRR12 pKa = 11.84TSRR15 pKa = 11.84PATVAVVVAALVTALLAGCDD35 pKa = 3.79APGKK39 pKa = 9.33EE40 pKa = 4.34QTPTLDD46 pKa = 3.55PSASSTQGADD56 pKa = 3.53PLAGAPAGYY65 pKa = 9.9EE66 pKa = 3.92SYY68 pKa = 11.01YY69 pKa = 10.88GQTLEE74 pKa = 4.39WEE76 pKa = 4.59SCGGGYY82 pKa = 10.3EE83 pKa = 4.38CTDD86 pKa = 3.41ADD88 pKa = 5.92APISWQDD95 pKa = 3.2PDD97 pKa = 4.26LGSIQLALKK106 pKa = 10.02RR107 pKa = 11.84LPASGDD113 pKa = 3.48RR114 pKa = 11.84QGSLLINPGGPGSSGVEE131 pKa = 4.14FVTSAASIIGDD142 pKa = 3.89RR143 pKa = 11.84VKK145 pKa = 10.79GAYY148 pKa = 9.88DD149 pKa = 3.44LVGFDD154 pKa = 3.75PRR156 pKa = 11.84GVGGSAPVKK165 pKa = 10.7CLDD168 pKa = 5.2DD169 pKa = 4.36ADD171 pKa = 4.6KK172 pKa = 11.17DD173 pKa = 3.88ASLSASYY180 pKa = 9.94PHH182 pKa = 5.69TTEE185 pKa = 5.62GIAAMADD192 pKa = 3.75DD193 pKa = 4.69LEE195 pKa = 5.26AWAQACDD202 pKa = 3.83EE203 pKa = 4.3NTGEE207 pKa = 4.37QLATVDD213 pKa = 3.81TQSAARR219 pKa = 11.84DD220 pKa = 3.49MDD222 pKa = 3.72MLRR225 pKa = 11.84AVLGDD230 pKa = 3.73EE231 pKa = 3.95QLHH234 pKa = 5.52YY235 pKa = 11.17LGFSYY240 pKa = 8.67GTQLGATYY248 pKa = 10.71AGLFPQTVGRR258 pKa = 11.84MVLDD262 pKa = 3.92GAIDD266 pKa = 3.58TTLDD270 pKa = 3.31ADD272 pKa = 4.53EE273 pKa = 5.19ISAEE277 pKa = 3.97QAVGFEE283 pKa = 4.25NALRR287 pKa = 11.84AYY289 pKa = 9.2VTDD292 pKa = 3.9CLGGRR297 pKa = 11.84ACPLSGSVDD306 pKa = 2.83GGMQQIRR313 pKa = 11.84DD314 pKa = 3.76LLDD317 pKa = 3.37RR318 pKa = 11.84TLEE321 pKa = 4.47DD322 pKa = 4.48PLPTDD327 pKa = 3.31SDD329 pKa = 3.86RR330 pKa = 11.84DD331 pKa = 3.7LTQSLTFYY339 pKa = 11.13GIALPLYY346 pKa = 10.64SKK348 pKa = 11.23ANWSLLTNALSDD360 pKa = 3.76ALEE363 pKa = 5.1DD364 pKa = 4.3GDD366 pKa = 5.63GSTLLYY372 pKa = 10.81LADD375 pKa = 5.28FYY377 pKa = 11.77NDD379 pKa = 3.72RR380 pKa = 11.84QPDD383 pKa = 3.58GSFSSNSAEE392 pKa = 3.77AFRR395 pKa = 11.84AINCLDD401 pKa = 3.5DD402 pKa = 5.18RR403 pKa = 11.84GNPDD407 pKa = 3.89PDD409 pKa = 3.9FMAEE413 pKa = 3.72QAEE416 pKa = 4.71EE417 pKa = 4.15IEE419 pKa = 4.39AAAPTMGEE427 pKa = 3.92FFTYY431 pKa = 10.4SGLTCADD438 pKa = 3.23WPVPEE443 pKa = 5.07VEE445 pKa = 4.37QQFDD449 pKa = 3.43LHH451 pKa = 7.65ASGAAPIVVVGTTNDD466 pKa = 3.55PATPYY471 pKa = 10.5VWAQGLAKK479 pKa = 9.67TLDD482 pKa = 3.54SATLVTFEE490 pKa = 4.97GEE492 pKa = 3.74GHH494 pKa = 5.44TAYY497 pKa = 10.31GSSNDD502 pKa = 3.9CVGDD506 pKa = 3.61AVDD509 pKa = 5.52DD510 pKa = 4.03YY511 pKa = 11.55LVDD514 pKa = 3.6GTVPKK519 pKa = 10.82DD520 pKa = 3.3GLTCC524 pKa = 4.55
Molecular weight: 54.98 kDa
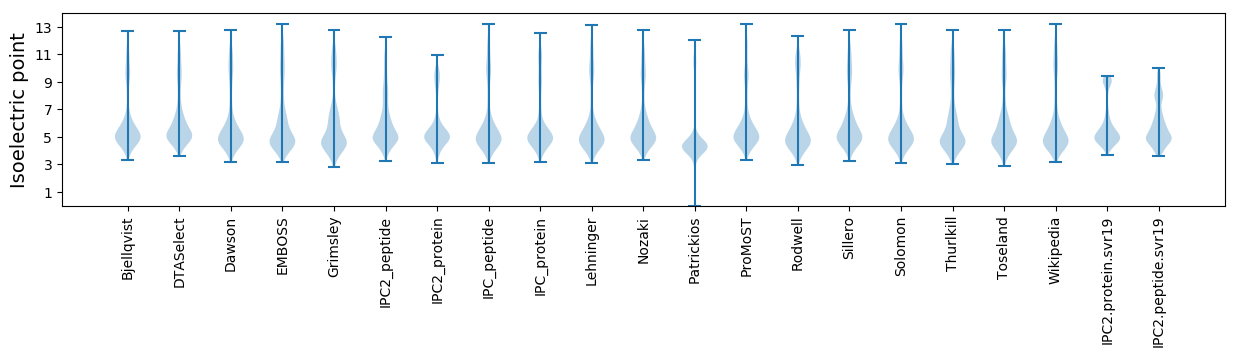
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9CCW4|A0A2M9CCW4_9CELL Uncharacterized protein OS=Sediminihabitans luteus OX=1138585 GN=CLV28_2695 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.61GRR40 pKa = 11.84AEE42 pKa = 3.84LSAA45 pKa = 4.93
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.61GRR40 pKa = 11.84AEE42 pKa = 3.84LSAA45 pKa = 4.93
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1036919 |
29 |
5544 |
346.2 |
36.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.55 ± 0.061 | 0.566 ± 0.01 |
6.909 ± 0.047 | 5.258 ± 0.042 |
2.495 ± 0.028 | 9.467 ± 0.045 |
2.081 ± 0.027 | 2.88 ± 0.032 |
1.519 ± 0.031 | 9.82 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.537 ± 0.019 | 1.487 ± 0.022 |
5.943 ± 0.047 | 2.464 ± 0.026 |
7.493 ± 0.061 | 5.092 ± 0.037 |
6.756 ± 0.069 | 10.388 ± 0.062 |
1.499 ± 0.019 | 1.796 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
