
Oerskovia sp. Root918
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Oerskovia; unclassified Oerskovia
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
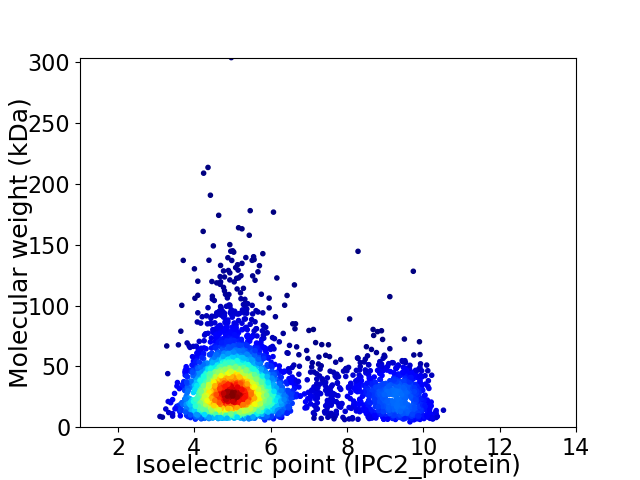
Virtual 2D-PAGE plot for 3618 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
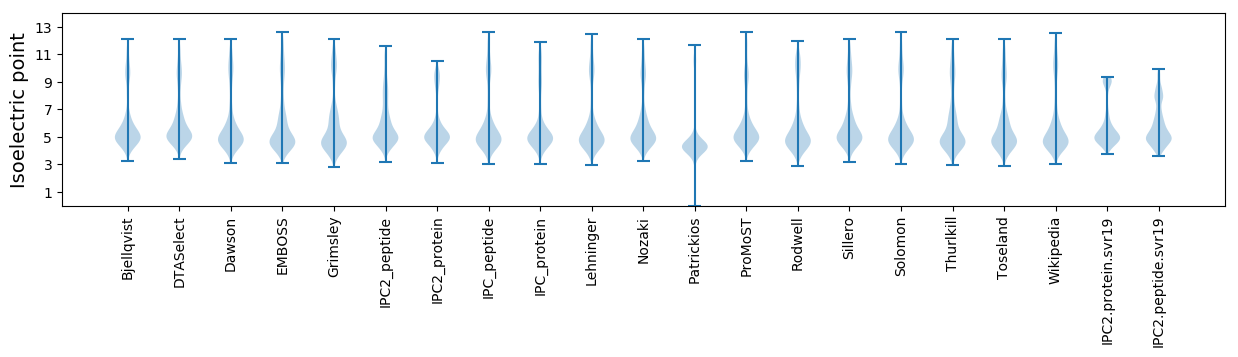
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q9C8V5|A0A0Q9C8V5_9CELL PDZ domain-containing protein OS=Oerskovia sp. Root918 OX=1736607 GN=ASE27_08870 PE=3 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84STLSIKK9 pKa = 10.17RR10 pKa = 11.84RR11 pKa = 11.84ALAAVAGGASLAIVLTACSGSGDD34 pKa = 3.79GGDD37 pKa = 4.64AGDD40 pKa = 4.35TEE42 pKa = 5.21AATADD47 pKa = 3.31CSAYY51 pKa = 10.13EE52 pKa = 4.12EE53 pKa = 4.45YY54 pKa = 11.32GDD56 pKa = 4.89LSGKK60 pKa = 7.19TVSVYY65 pKa = 7.36TTIVDD70 pKa = 4.0TEE72 pKa = 4.42AEE74 pKa = 4.2QQQEE78 pKa = 4.38SYY80 pKa = 11.2KK81 pKa = 10.7PFEE84 pKa = 5.16DD85 pKa = 3.83CTGATIKK92 pKa = 11.22YY93 pKa = 8.16EE94 pKa = 3.98GSKK97 pKa = 10.36EE98 pKa = 3.94FEE100 pKa = 3.94AQLPIRR106 pKa = 11.84IQSGSAPDD114 pKa = 3.29IAYY117 pKa = 9.39IPQPGFLKK125 pKa = 10.82SLVQDD130 pKa = 3.77YY131 pKa = 10.44PDD133 pKa = 5.3AILPVGDD140 pKa = 3.71AASANVDD147 pKa = 3.4EE148 pKa = 6.18FYY150 pKa = 10.83TEE152 pKa = 3.38SWRR155 pKa = 11.84DD156 pKa = 3.39YY157 pKa = 9.35GTVDD161 pKa = 2.94GTLYY165 pKa = 9.35ATPLGANVKK174 pKa = 10.15SFVWYY179 pKa = 9.88SPAMFEE185 pKa = 4.41DD186 pKa = 3.58AGYY189 pKa = 9.6EE190 pKa = 4.43IPTTWDD196 pKa = 3.11EE197 pKa = 5.09LIALSDD203 pKa = 4.31QIVEE207 pKa = 4.52DD208 pKa = 4.27NPDD211 pKa = 3.5GEE213 pKa = 4.42IKK215 pKa = 9.93PWCAGIGSGEE225 pKa = 4.01ATGWPATDD233 pKa = 2.82WMEE236 pKa = 4.54DD237 pKa = 3.36VVLRR241 pKa = 11.84TAGPEE246 pKa = 4.65VYY248 pKa = 9.99DD249 pKa = 2.9QWVNHH254 pKa = 6.96EE255 pKa = 4.64IPFNDD260 pKa = 3.7PQIAEE265 pKa = 4.15ALATVGTVLKK275 pKa = 10.89NDD277 pKa = 4.02DD278 pKa = 4.18YY279 pKa = 11.76VNGGIGDD286 pKa = 3.76VSSIATTEE294 pKa = 4.01FTEE297 pKa = 4.68GGLPILDD304 pKa = 4.05GLCYY308 pKa = 8.61MHH310 pKa = 6.93RR311 pKa = 11.84QASFYY316 pKa = 9.03QANWPEE322 pKa = 4.18GTTVAEE328 pKa = 4.93DD329 pKa = 3.73GDD331 pKa = 3.96VFAFYY336 pKa = 10.51FPGNDD341 pKa = 3.32TEE343 pKa = 4.54AKK345 pKa = 8.76PVLGGGEE352 pKa = 4.21FVVSFADD359 pKa = 3.87RR360 pKa = 11.84PEE362 pKa = 4.05VQAFQAFLTSPEE374 pKa = 3.78WSNAKK379 pKa = 8.46ATATPQGWISANKK392 pKa = 9.91EE393 pKa = 3.7LDD395 pKa = 3.27KK396 pKa = 11.47SLLQSPIDD404 pKa = 3.55QLSYY408 pKa = 11.67GLLTDD413 pKa = 3.9DD414 pKa = 4.27SYY416 pKa = 12.3VFRR419 pKa = 11.84FDD421 pKa = 4.05GSDD424 pKa = 3.31MMPAAVGAGSFWTEE438 pKa = 3.47MTEE441 pKa = 4.34WIASDD446 pKa = 3.56KK447 pKa = 11.3SDD449 pKa = 3.65TAVLDD454 pKa = 4.8AIEE457 pKa = 4.77ASWPTSS463 pKa = 3.11
MM1 pKa = 7.42ARR3 pKa = 11.84STLSIKK9 pKa = 10.17RR10 pKa = 11.84RR11 pKa = 11.84ALAAVAGGASLAIVLTACSGSGDD34 pKa = 3.79GGDD37 pKa = 4.64AGDD40 pKa = 4.35TEE42 pKa = 5.21AATADD47 pKa = 3.31CSAYY51 pKa = 10.13EE52 pKa = 4.12EE53 pKa = 4.45YY54 pKa = 11.32GDD56 pKa = 4.89LSGKK60 pKa = 7.19TVSVYY65 pKa = 7.36TTIVDD70 pKa = 4.0TEE72 pKa = 4.42AEE74 pKa = 4.2QQQEE78 pKa = 4.38SYY80 pKa = 11.2KK81 pKa = 10.7PFEE84 pKa = 5.16DD85 pKa = 3.83CTGATIKK92 pKa = 11.22YY93 pKa = 8.16EE94 pKa = 3.98GSKK97 pKa = 10.36EE98 pKa = 3.94FEE100 pKa = 3.94AQLPIRR106 pKa = 11.84IQSGSAPDD114 pKa = 3.29IAYY117 pKa = 9.39IPQPGFLKK125 pKa = 10.82SLVQDD130 pKa = 3.77YY131 pKa = 10.44PDD133 pKa = 5.3AILPVGDD140 pKa = 3.71AASANVDD147 pKa = 3.4EE148 pKa = 6.18FYY150 pKa = 10.83TEE152 pKa = 3.38SWRR155 pKa = 11.84DD156 pKa = 3.39YY157 pKa = 9.35GTVDD161 pKa = 2.94GTLYY165 pKa = 9.35ATPLGANVKK174 pKa = 10.15SFVWYY179 pKa = 9.88SPAMFEE185 pKa = 4.41DD186 pKa = 3.58AGYY189 pKa = 9.6EE190 pKa = 4.43IPTTWDD196 pKa = 3.11EE197 pKa = 5.09LIALSDD203 pKa = 4.31QIVEE207 pKa = 4.52DD208 pKa = 4.27NPDD211 pKa = 3.5GEE213 pKa = 4.42IKK215 pKa = 9.93PWCAGIGSGEE225 pKa = 4.01ATGWPATDD233 pKa = 2.82WMEE236 pKa = 4.54DD237 pKa = 3.36VVLRR241 pKa = 11.84TAGPEE246 pKa = 4.65VYY248 pKa = 9.99DD249 pKa = 2.9QWVNHH254 pKa = 6.96EE255 pKa = 4.64IPFNDD260 pKa = 3.7PQIAEE265 pKa = 4.15ALATVGTVLKK275 pKa = 10.89NDD277 pKa = 4.02DD278 pKa = 4.18YY279 pKa = 11.76VNGGIGDD286 pKa = 3.76VSSIATTEE294 pKa = 4.01FTEE297 pKa = 4.68GGLPILDD304 pKa = 4.05GLCYY308 pKa = 8.61MHH310 pKa = 6.93RR311 pKa = 11.84QASFYY316 pKa = 9.03QANWPEE322 pKa = 4.18GTTVAEE328 pKa = 4.93DD329 pKa = 3.73GDD331 pKa = 3.96VFAFYY336 pKa = 10.51FPGNDD341 pKa = 3.32TEE343 pKa = 4.54AKK345 pKa = 8.76PVLGGGEE352 pKa = 4.21FVVSFADD359 pKa = 3.87RR360 pKa = 11.84PEE362 pKa = 4.05VQAFQAFLTSPEE374 pKa = 3.78WSNAKK379 pKa = 8.46ATATPQGWISANKK392 pKa = 9.91EE393 pKa = 3.7LDD395 pKa = 3.27KK396 pKa = 11.47SLLQSPIDD404 pKa = 3.55QLSYY408 pKa = 11.67GLLTDD413 pKa = 3.9DD414 pKa = 4.27SYY416 pKa = 12.3VFRR419 pKa = 11.84FDD421 pKa = 4.05GSDD424 pKa = 3.31MMPAAVGAGSFWTEE438 pKa = 3.47MTEE441 pKa = 4.34WIASDD446 pKa = 3.56KK447 pKa = 11.3SDD449 pKa = 3.65TAVLDD454 pKa = 4.8AIEE457 pKa = 4.77ASWPTSS463 pKa = 3.11
Molecular weight: 49.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q9C1Z8|A0A0Q9C1Z8_9CELL MarR family transcriptional regulator OS=Oerskovia sp. Root918 OX=1736607 GN=ASE27_07240 PE=4 SV=1
MM1 pKa = 7.49AGEE4 pKa = 3.79WTIRR8 pKa = 11.84EE9 pKa = 4.37LARR12 pKa = 11.84KK13 pKa = 9.6AGVSRR18 pKa = 11.84KK19 pKa = 9.32SVWAWIDD26 pKa = 3.28AQGWARR32 pKa = 11.84PVSGPWILDD41 pKa = 3.25GEE43 pKa = 4.42RR44 pKa = 11.84ARR46 pKa = 11.84LVLEE50 pKa = 4.58RR51 pKa = 11.84FEE53 pKa = 4.03QTAPLRR59 pKa = 11.84TPRR62 pKa = 11.84EE63 pKa = 3.94PVPCSIEE70 pKa = 3.85GCEE73 pKa = 3.85RR74 pKa = 11.84TRR76 pKa = 11.84AGLQDD81 pKa = 3.93MCKK84 pKa = 8.57MHH86 pKa = 5.4YY87 pKa = 8.42QRR89 pKa = 11.84RR90 pKa = 11.84LRR92 pKa = 11.84TGRR95 pKa = 11.84TEE97 pKa = 3.93RR98 pKa = 11.84SSGGDD103 pKa = 2.79WQTAKK108 pKa = 8.47THH110 pKa = 6.29CPAGHH115 pKa = 6.9EE116 pKa = 3.95YY117 pKa = 10.35RR118 pKa = 11.84PEE120 pKa = 4.09NIYY123 pKa = 10.56RR124 pKa = 11.84FPSDD128 pKa = 2.45VGTRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84CRR136 pKa = 11.84TCRR139 pKa = 11.84IAQSSVSKK147 pKa = 10.79KK148 pKa = 10.22PSS150 pKa = 2.95
MM1 pKa = 7.49AGEE4 pKa = 3.79WTIRR8 pKa = 11.84EE9 pKa = 4.37LARR12 pKa = 11.84KK13 pKa = 9.6AGVSRR18 pKa = 11.84KK19 pKa = 9.32SVWAWIDD26 pKa = 3.28AQGWARR32 pKa = 11.84PVSGPWILDD41 pKa = 3.25GEE43 pKa = 4.42RR44 pKa = 11.84ARR46 pKa = 11.84LVLEE50 pKa = 4.58RR51 pKa = 11.84FEE53 pKa = 4.03QTAPLRR59 pKa = 11.84TPRR62 pKa = 11.84EE63 pKa = 3.94PVPCSIEE70 pKa = 3.85GCEE73 pKa = 3.85RR74 pKa = 11.84TRR76 pKa = 11.84AGLQDD81 pKa = 3.93MCKK84 pKa = 8.57MHH86 pKa = 5.4YY87 pKa = 8.42QRR89 pKa = 11.84RR90 pKa = 11.84LRR92 pKa = 11.84TGRR95 pKa = 11.84TEE97 pKa = 3.93RR98 pKa = 11.84SSGGDD103 pKa = 2.79WQTAKK108 pKa = 8.47THH110 pKa = 6.29CPAGHH115 pKa = 6.9EE116 pKa = 3.95YY117 pKa = 10.35RR118 pKa = 11.84PEE120 pKa = 4.09NIYY123 pKa = 10.56RR124 pKa = 11.84FPSDD128 pKa = 2.45VGTRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84CRR136 pKa = 11.84TCRR139 pKa = 11.84IAQSSVSKK147 pKa = 10.79KK148 pKa = 10.22PSS150 pKa = 2.95
Molecular weight: 17.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1211724 |
37 |
2957 |
334.9 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.817 ± 0.063 | 0.539 ± 0.009 |
6.275 ± 0.034 | 5.576 ± 0.038 |
2.733 ± 0.026 | 9.482 ± 0.039 |
2.0 ± 0.021 | 3.292 ± 0.035 |
1.691 ± 0.03 | 10.238 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.599 ± 0.016 | 1.654 ± 0.023 |
5.876 ± 0.034 | 2.701 ± 0.021 |
7.211 ± 0.047 | 5.503 ± 0.029 |
6.464 ± 0.041 | 9.934 ± 0.046 |
1.526 ± 0.016 | 1.887 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
