
Bettongia penicillata papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyolambdapapillomavirus; Dyolambdapapillomavirus 1
Average proteome isoelectric point is 5.46
Get precalculated fractions of proteins
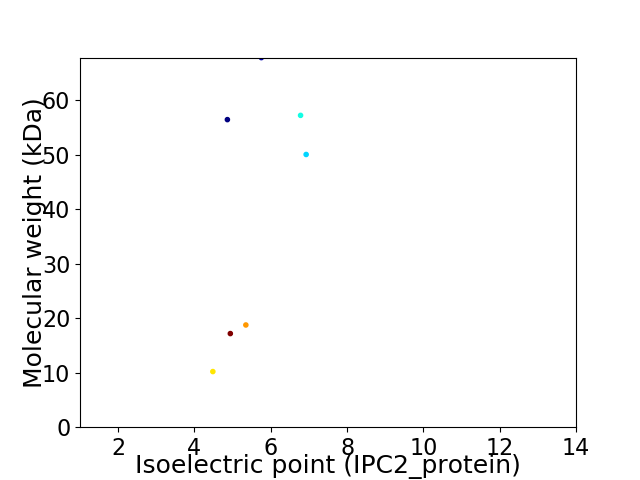
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6N1C0|D6N1C0_9PAPI Replication protein E1 OS=Bettongia penicillata papillomavirus 1 OX=759701 GN=E1 PE=3 SV=1
MM1 pKa = 7.76IGDD4 pKa = 5.04RR5 pKa = 11.84IDD7 pKa = 4.05LKK9 pKa = 11.27DD10 pKa = 3.58IVLQLDD16 pKa = 3.62QVVPTSEE23 pKa = 4.33EE24 pKa = 3.87EE25 pKa = 4.16STTGDD30 pKa = 3.5EE31 pKa = 4.7VEE33 pKa = 4.16HH34 pKa = 6.7RR35 pKa = 11.84PYY37 pKa = 10.71KK38 pKa = 10.56YY39 pKa = 10.31YY40 pKa = 10.95DD41 pKa = 3.51VTVSCATCSRR51 pKa = 11.84RR52 pKa = 11.84ISFCILANPEE62 pKa = 4.17TVSGFFNYY70 pKa = 9.47FFEE73 pKa = 4.78DD74 pKa = 3.46VRR76 pKa = 11.84PVCLPCVQKK85 pKa = 11.35GEE87 pKa = 4.14NGQQ90 pKa = 3.41
MM1 pKa = 7.76IGDD4 pKa = 5.04RR5 pKa = 11.84IDD7 pKa = 4.05LKK9 pKa = 11.27DD10 pKa = 3.58IVLQLDD16 pKa = 3.62QVVPTSEE23 pKa = 4.33EE24 pKa = 3.87EE25 pKa = 4.16STTGDD30 pKa = 3.5EE31 pKa = 4.7VEE33 pKa = 4.16HH34 pKa = 6.7RR35 pKa = 11.84PYY37 pKa = 10.71KK38 pKa = 10.56YY39 pKa = 10.31YY40 pKa = 10.95DD41 pKa = 3.51VTVSCATCSRR51 pKa = 11.84RR52 pKa = 11.84ISFCILANPEE62 pKa = 4.17TVSGFFNYY70 pKa = 9.47FFEE73 pKa = 4.78DD74 pKa = 3.46VRR76 pKa = 11.84PVCLPCVQKK85 pKa = 11.35GEE87 pKa = 4.14NGQQ90 pKa = 3.41
Molecular weight: 10.21 kDa
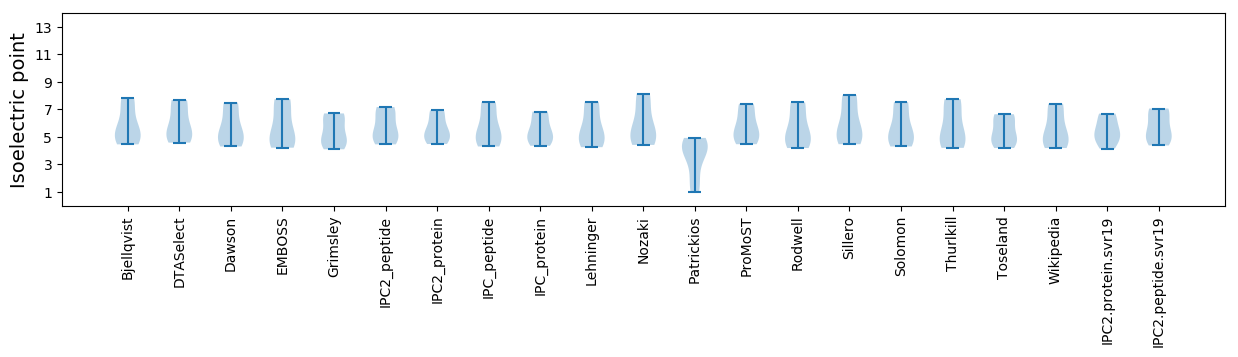
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6N1C2|D6N1C2_9PAPI E4 (Fragment) OS=Bettongia penicillata papillomavirus 1 OX=759701 PE=4 SV=1
MM1 pKa = 7.0EE2 pKa = 4.65TLTARR7 pKa = 11.84FDD9 pKa = 3.75AVQGKK14 pKa = 9.99LMDD17 pKa = 4.59LFEE20 pKa = 6.07KK21 pKa = 10.64DD22 pKa = 3.25SNDD25 pKa = 3.33IVDD28 pKa = 3.65HH29 pKa = 6.28CEE31 pKa = 3.5YY32 pKa = 10.28WSLIRR37 pKa = 11.84QEE39 pKa = 4.38NIMMNFGRR47 pKa = 11.84KK48 pKa = 9.09KK49 pKa = 9.4GLKK52 pKa = 9.44TIGLQPLPLSAVSEE66 pKa = 4.32AQAKK70 pKa = 9.31IAIEE74 pKa = 3.92MEE76 pKa = 4.39LVVKK80 pKa = 8.06TLKK83 pKa = 10.57NSPFGDD89 pKa = 4.04EE90 pKa = 4.23KK91 pKa = 10.29WTFSDD96 pKa = 3.35TSYY99 pKa = 9.58EE100 pKa = 4.43TYY102 pKa = 10.45VSPPQRR108 pKa = 11.84CFKK111 pKa = 10.4KK112 pKa = 10.51LPYY115 pKa = 9.67LVKK118 pKa = 10.33IKK120 pKa = 10.46HH121 pKa = 6.13GPNMDD126 pKa = 3.53NVDD129 pKa = 3.93VYY131 pKa = 10.53TGWGKK136 pKa = 10.1IYY138 pKa = 10.61KK139 pKa = 9.39QDD141 pKa = 3.84CQGEE145 pKa = 4.44WTVVTGEE152 pKa = 4.17TDD154 pKa = 3.06EE155 pKa = 4.51TGCFYY160 pKa = 11.38VEE162 pKa = 4.95DD163 pKa = 4.09DD164 pKa = 3.77CKK166 pKa = 11.17VYY168 pKa = 11.25YY169 pKa = 10.29KK170 pKa = 11.01VFDD173 pKa = 4.19IDD175 pKa = 3.54SHH177 pKa = 7.82NYY179 pKa = 10.86DD180 pKa = 3.38MTFKK184 pKa = 10.91HH185 pKa = 6.38PLNSDD190 pKa = 4.14FVRR193 pKa = 11.84KK194 pKa = 5.96THH196 pKa = 6.9RR197 pKa = 11.84GRR199 pKa = 11.84GQGEE203 pKa = 4.1QDD205 pKa = 3.7STPQTPSSQPEE216 pKa = 3.77RR217 pKa = 11.84TKK219 pKa = 10.73EE220 pKa = 3.86RR221 pKa = 11.84KK222 pKa = 7.15TGPRR226 pKa = 11.84VRR228 pKa = 11.84TSGTYY233 pKa = 8.47TPPVGPLGGAQPIPSSAPKK252 pKa = 8.32EE253 pKa = 4.67TPDD256 pKa = 3.55GAHH259 pKa = 7.02PGGGGVPRR267 pKa = 11.84GVGGRR272 pKa = 11.84CDD274 pKa = 3.42QPGGSGGPRR283 pKa = 11.84KK284 pKa = 9.68RR285 pKa = 11.84AWEE288 pKa = 3.93EE289 pKa = 3.77GGGGGEE295 pKa = 4.19EE296 pKa = 4.5EE297 pKa = 4.36EE298 pKa = 5.02GEE300 pKa = 4.46APRR303 pKa = 11.84TSKK306 pKa = 10.73KK307 pKa = 9.91PAGGTDD313 pKa = 3.26RR314 pKa = 11.84PLEE317 pKa = 4.34GQTGGRR323 pKa = 11.84PGEE326 pKa = 4.34VTSTTQRR333 pKa = 11.84DD334 pKa = 3.91CRR336 pKa = 11.84EE337 pKa = 4.44GPSSCPVTCPVTCPEE352 pKa = 4.17TSCPVTSCPVTSPEE366 pKa = 4.17TQCPVTQCPVLLAQGDD382 pKa = 4.23ANILKK387 pKa = 9.31CWRR390 pKa = 11.84YY391 pKa = 9.91RR392 pKa = 11.84LEE394 pKa = 4.29TKK396 pKa = 10.41HH397 pKa = 6.45KK398 pKa = 9.12STYY401 pKa = 10.22KK402 pKa = 10.28HH403 pKa = 6.83ISSTWYY409 pKa = 5.34WTAGGRR415 pKa = 11.84GQGKK419 pKa = 8.64HH420 pKa = 5.54RR421 pKa = 11.84MLIAFDD427 pKa = 3.82SAQQEE432 pKa = 4.26KK433 pKa = 11.21DD434 pKa = 3.41FLLLKK439 pKa = 10.1LPKK442 pKa = 10.33GVFVSKK448 pKa = 10.55GWFQNLL454 pKa = 3.01
MM1 pKa = 7.0EE2 pKa = 4.65TLTARR7 pKa = 11.84FDD9 pKa = 3.75AVQGKK14 pKa = 9.99LMDD17 pKa = 4.59LFEE20 pKa = 6.07KK21 pKa = 10.64DD22 pKa = 3.25SNDD25 pKa = 3.33IVDD28 pKa = 3.65HH29 pKa = 6.28CEE31 pKa = 3.5YY32 pKa = 10.28WSLIRR37 pKa = 11.84QEE39 pKa = 4.38NIMMNFGRR47 pKa = 11.84KK48 pKa = 9.09KK49 pKa = 9.4GLKK52 pKa = 9.44TIGLQPLPLSAVSEE66 pKa = 4.32AQAKK70 pKa = 9.31IAIEE74 pKa = 3.92MEE76 pKa = 4.39LVVKK80 pKa = 8.06TLKK83 pKa = 10.57NSPFGDD89 pKa = 4.04EE90 pKa = 4.23KK91 pKa = 10.29WTFSDD96 pKa = 3.35TSYY99 pKa = 9.58EE100 pKa = 4.43TYY102 pKa = 10.45VSPPQRR108 pKa = 11.84CFKK111 pKa = 10.4KK112 pKa = 10.51LPYY115 pKa = 9.67LVKK118 pKa = 10.33IKK120 pKa = 10.46HH121 pKa = 6.13GPNMDD126 pKa = 3.53NVDD129 pKa = 3.93VYY131 pKa = 10.53TGWGKK136 pKa = 10.1IYY138 pKa = 10.61KK139 pKa = 9.39QDD141 pKa = 3.84CQGEE145 pKa = 4.44WTVVTGEE152 pKa = 4.17TDD154 pKa = 3.06EE155 pKa = 4.51TGCFYY160 pKa = 11.38VEE162 pKa = 4.95DD163 pKa = 4.09DD164 pKa = 3.77CKK166 pKa = 11.17VYY168 pKa = 11.25YY169 pKa = 10.29KK170 pKa = 11.01VFDD173 pKa = 4.19IDD175 pKa = 3.54SHH177 pKa = 7.82NYY179 pKa = 10.86DD180 pKa = 3.38MTFKK184 pKa = 10.91HH185 pKa = 6.38PLNSDD190 pKa = 4.14FVRR193 pKa = 11.84KK194 pKa = 5.96THH196 pKa = 6.9RR197 pKa = 11.84GRR199 pKa = 11.84GQGEE203 pKa = 4.1QDD205 pKa = 3.7STPQTPSSQPEE216 pKa = 3.77RR217 pKa = 11.84TKK219 pKa = 10.73EE220 pKa = 3.86RR221 pKa = 11.84KK222 pKa = 7.15TGPRR226 pKa = 11.84VRR228 pKa = 11.84TSGTYY233 pKa = 8.47TPPVGPLGGAQPIPSSAPKK252 pKa = 8.32EE253 pKa = 4.67TPDD256 pKa = 3.55GAHH259 pKa = 7.02PGGGGVPRR267 pKa = 11.84GVGGRR272 pKa = 11.84CDD274 pKa = 3.42QPGGSGGPRR283 pKa = 11.84KK284 pKa = 9.68RR285 pKa = 11.84AWEE288 pKa = 3.93EE289 pKa = 3.77GGGGGEE295 pKa = 4.19EE296 pKa = 4.5EE297 pKa = 4.36EE298 pKa = 5.02GEE300 pKa = 4.46APRR303 pKa = 11.84TSKK306 pKa = 10.73KK307 pKa = 9.91PAGGTDD313 pKa = 3.26RR314 pKa = 11.84PLEE317 pKa = 4.34GQTGGRR323 pKa = 11.84PGEE326 pKa = 4.34VTSTTQRR333 pKa = 11.84DD334 pKa = 3.91CRR336 pKa = 11.84EE337 pKa = 4.44GPSSCPVTCPVTCPEE352 pKa = 4.17TSCPVTSCPVTSPEE366 pKa = 4.17TQCPVTQCPVLLAQGDD382 pKa = 4.23ANILKK387 pKa = 9.31CWRR390 pKa = 11.84YY391 pKa = 9.91RR392 pKa = 11.84LEE394 pKa = 4.29TKK396 pKa = 10.41HH397 pKa = 6.45KK398 pKa = 9.12STYY401 pKa = 10.22KK402 pKa = 10.28HH403 pKa = 6.83ISSTWYY409 pKa = 5.34WTAGGRR415 pKa = 11.84GQGKK419 pKa = 8.64HH420 pKa = 5.54RR421 pKa = 11.84MLIAFDD427 pKa = 3.82SAQQEE432 pKa = 4.26KK433 pKa = 11.21DD434 pKa = 3.41FLLLKK439 pKa = 10.1LPKK442 pKa = 10.33GVFVSKK448 pKa = 10.55GWFQNLL454 pKa = 3.01
Molecular weight: 50.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2475 |
90 |
596 |
353.6 |
39.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.485 ± 0.466 | 2.626 ± 0.804 |
6.545 ± 0.265 | 7.111 ± 0.729 |
4.485 ± 0.443 | 7.232 ± 1.012 |
1.778 ± 0.151 | 4.121 ± 0.261 |
5.98 ± 0.94 | 7.879 ± 0.887 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.374 ± 0.293 | 4.121 ± 0.825 |
7.798 ± 1.375 | 3.96 ± 0.433 |
5.939 ± 0.623 | 7.273 ± 0.651 |
6.263 ± 0.562 | 6.626 ± 0.643 |
1.455 ± 0.289 | 2.949 ± 0.437 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
