
Hyphomicrobium sp. xq
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Hyphomicrobiaceae; Hyphomicrobium; unclassified Hyphomicrobium
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
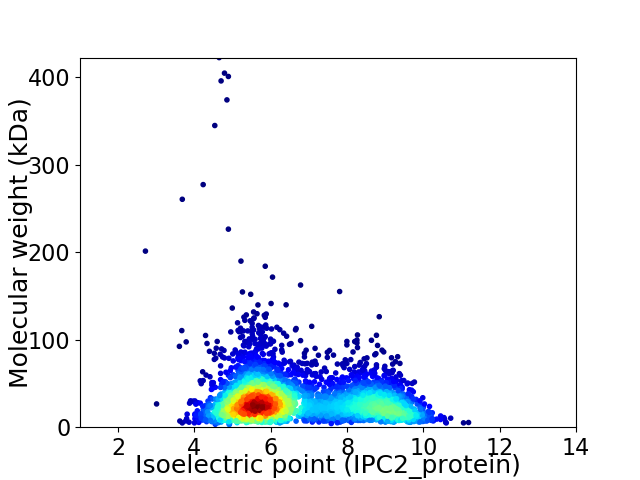
Virtual 2D-PAGE plot for 3664 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
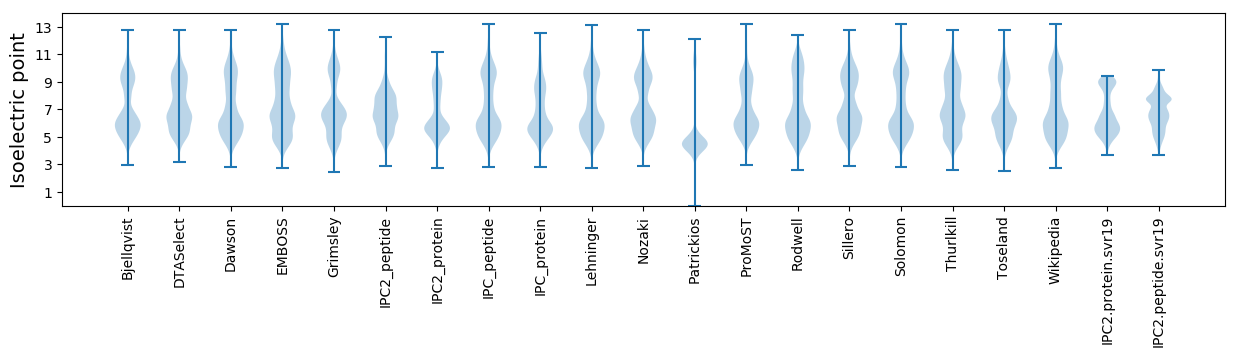
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I3KGD7|A0A6I3KGD7_9RHIZ M48 family metalloprotease OS=Hyphomicrobium sp. xq OX=2665159 GN=GIW81_06545 PE=4 SV=1
MM1 pKa = 7.88WLTALAVVDD10 pKa = 4.3VGPAHH15 pKa = 6.63AQAITPPSPPEE26 pKa = 4.1CVSVTGIGVEE36 pKa = 4.8IITCTGNLSPGVNLANGPGPYY57 pKa = 9.83DD58 pKa = 3.25ILNVHH63 pKa = 6.44SLTANITPASGTAGILFTSDD83 pKa = 2.73GSVEE87 pKa = 4.35LNVDD91 pKa = 3.89TGPFGIFTTAADD103 pKa = 4.18GVFAAGDD110 pKa = 3.89AGPVTINFFGAITTSGNGAVGIRR133 pKa = 11.84AGGLDD138 pKa = 3.84DD139 pKa = 4.97LVSIVASGTIVTDD152 pKa = 3.75GLSASGIAASTDD164 pKa = 3.05NGPIDD169 pKa = 3.42IRR171 pKa = 11.84SSVSIATQGQSAHH184 pKa = 6.69GIDD187 pKa = 3.76VSSVGGAINILSMGNIATGGTDD209 pKa = 3.95AIGIYY214 pKa = 8.9ATSTTGQITITSTSDD229 pKa = 2.41ITTSGGDD236 pKa = 3.33SVGIFAEE243 pKa = 4.41TSGVAMVEE251 pKa = 4.03SSGNFLTRR259 pKa = 11.84GDD261 pKa = 3.78GAAGIAAAGQTGTAVISTGSIATEE285 pKa = 4.33GSDD288 pKa = 4.2APGITAISDD297 pKa = 3.85AEE299 pKa = 4.33VGVAMSGSIATARR312 pKa = 11.84DD313 pKa = 3.25NSRR316 pKa = 11.84GIWAQSLVNGMVAVGAGGTITTAGDD341 pKa = 3.54DD342 pKa = 4.47SDD344 pKa = 6.08GIWADD349 pKa = 3.46SSSGAVVVISSANITTTGQQSDD371 pKa = 4.77GISAISGAALATVVNFGVVSANGVGSAGIFAEE403 pKa = 4.57GQEE406 pKa = 4.54TQVEE410 pKa = 4.63NYY412 pKa = 7.93GTVAGGPCCGGVMMDD427 pKa = 4.04AVDD430 pKa = 4.4LAVLMNYY437 pKa = 7.76GTIIGQTSGEE447 pKa = 4.32AIFGSGTDD455 pKa = 3.37VRR457 pKa = 11.84IEE459 pKa = 3.93NFGTVTGNVILFGSNSSYY477 pKa = 10.9FFNQAGALFNSGEE490 pKa = 4.09QVMSGFVVNDD500 pKa = 3.51GTIAPGGRR508 pKa = 11.84GSVLTTLMSDD518 pKa = 5.11FYY520 pKa = 11.35SQGAGGTYY528 pKa = 10.47AVDD531 pKa = 5.16LDD533 pKa = 4.21PQASNPSDD541 pKa = 3.58RR542 pKa = 11.84NDD544 pKa = 3.66YY545 pKa = 10.53IVASNNAEE553 pKa = 3.91VAGVVEE559 pKa = 4.52VSVLSLPVAASEE571 pKa = 4.31TFIILTGMSSLTDD584 pKa = 3.29NGLSLIASPALHH596 pKa = 6.08ATLFTDD602 pKa = 3.97ATNVTLGISVDD613 pKa = 4.19FTVDD617 pKa = 3.17SLNPNQRR624 pKa = 11.84HH625 pKa = 5.38IAADD629 pKa = 4.02LNQVFLAGGGGVTPVLLGLLNTGNFDD655 pKa = 3.75AYY657 pKa = 10.15KK658 pKa = 10.58NALNQLSPEE667 pKa = 4.09IYY669 pKa = 10.52SNAEE673 pKa = 3.12IAALYY678 pKa = 10.62SSLAFSNSLLSCKK691 pKa = 10.5VNGTGTGTAAIIRR704 pKa = 11.84EE705 pKa = 4.68GQCLWAGASANFVDD719 pKa = 4.82TGTTSDD725 pKa = 4.03QIGFTQTTGLFNAGAQVALDD745 pKa = 3.85DD746 pKa = 3.66VWRR749 pKa = 11.84VGFGAGYY756 pKa = 10.01QSSSLQTATNAQSEE770 pKa = 4.73GSLAQAGLSVKK781 pKa = 10.23YY782 pKa = 10.83NPGPLLLAGAVSGGGAWYY800 pKa = 7.87DD801 pKa = 3.63TTRR804 pKa = 11.84TMAFGGFTGVAEE816 pKa = 4.56GDD818 pKa = 3.3QDD820 pKa = 3.78VGIVSGSLRR829 pKa = 11.84AAYY832 pKa = 10.32VLGSPHH838 pKa = 7.45LYY840 pKa = 10.07YY841 pKa = 10.72KK842 pKa = 10.61PILDD846 pKa = 4.5LGLTHH851 pKa = 7.27LEE853 pKa = 3.9LGGFTEE859 pKa = 4.57SGGGGAALTVAGGGQTVFSLAPTLEE884 pKa = 4.4VGSEE888 pKa = 3.26WWMSNGTLIRR898 pKa = 11.84PMIRR902 pKa = 11.84GGAIWYY908 pKa = 9.4DD909 pKa = 3.38GADD912 pKa = 3.68FALTAAFAGAPLGVSPFTINTNIDD936 pKa = 3.26EE937 pKa = 4.35VMGLVGAGVEE947 pKa = 4.46VINGGDD953 pKa = 3.07AALRR957 pKa = 11.84FSYY960 pKa = 10.38DD961 pKa = 3.09GQLGEE966 pKa = 4.3TTQIHH971 pKa = 4.78TVGVKK976 pKa = 10.14GSARR980 pKa = 11.84FF981 pKa = 3.39
MM1 pKa = 7.88WLTALAVVDD10 pKa = 4.3VGPAHH15 pKa = 6.63AQAITPPSPPEE26 pKa = 4.1CVSVTGIGVEE36 pKa = 4.8IITCTGNLSPGVNLANGPGPYY57 pKa = 9.83DD58 pKa = 3.25ILNVHH63 pKa = 6.44SLTANITPASGTAGILFTSDD83 pKa = 2.73GSVEE87 pKa = 4.35LNVDD91 pKa = 3.89TGPFGIFTTAADD103 pKa = 4.18GVFAAGDD110 pKa = 3.89AGPVTINFFGAITTSGNGAVGIRR133 pKa = 11.84AGGLDD138 pKa = 3.84DD139 pKa = 4.97LVSIVASGTIVTDD152 pKa = 3.75GLSASGIAASTDD164 pKa = 3.05NGPIDD169 pKa = 3.42IRR171 pKa = 11.84SSVSIATQGQSAHH184 pKa = 6.69GIDD187 pKa = 3.76VSSVGGAINILSMGNIATGGTDD209 pKa = 3.95AIGIYY214 pKa = 8.9ATSTTGQITITSTSDD229 pKa = 2.41ITTSGGDD236 pKa = 3.33SVGIFAEE243 pKa = 4.41TSGVAMVEE251 pKa = 4.03SSGNFLTRR259 pKa = 11.84GDD261 pKa = 3.78GAAGIAAAGQTGTAVISTGSIATEE285 pKa = 4.33GSDD288 pKa = 4.2APGITAISDD297 pKa = 3.85AEE299 pKa = 4.33VGVAMSGSIATARR312 pKa = 11.84DD313 pKa = 3.25NSRR316 pKa = 11.84GIWAQSLVNGMVAVGAGGTITTAGDD341 pKa = 3.54DD342 pKa = 4.47SDD344 pKa = 6.08GIWADD349 pKa = 3.46SSSGAVVVISSANITTTGQQSDD371 pKa = 4.77GISAISGAALATVVNFGVVSANGVGSAGIFAEE403 pKa = 4.57GQEE406 pKa = 4.54TQVEE410 pKa = 4.63NYY412 pKa = 7.93GTVAGGPCCGGVMMDD427 pKa = 4.04AVDD430 pKa = 4.4LAVLMNYY437 pKa = 7.76GTIIGQTSGEE447 pKa = 4.32AIFGSGTDD455 pKa = 3.37VRR457 pKa = 11.84IEE459 pKa = 3.93NFGTVTGNVILFGSNSSYY477 pKa = 10.9FFNQAGALFNSGEE490 pKa = 4.09QVMSGFVVNDD500 pKa = 3.51GTIAPGGRR508 pKa = 11.84GSVLTTLMSDD518 pKa = 5.11FYY520 pKa = 11.35SQGAGGTYY528 pKa = 10.47AVDD531 pKa = 5.16LDD533 pKa = 4.21PQASNPSDD541 pKa = 3.58RR542 pKa = 11.84NDD544 pKa = 3.66YY545 pKa = 10.53IVASNNAEE553 pKa = 3.91VAGVVEE559 pKa = 4.52VSVLSLPVAASEE571 pKa = 4.31TFIILTGMSSLTDD584 pKa = 3.29NGLSLIASPALHH596 pKa = 6.08ATLFTDD602 pKa = 3.97ATNVTLGISVDD613 pKa = 4.19FTVDD617 pKa = 3.17SLNPNQRR624 pKa = 11.84HH625 pKa = 5.38IAADD629 pKa = 4.02LNQVFLAGGGGVTPVLLGLLNTGNFDD655 pKa = 3.75AYY657 pKa = 10.15KK658 pKa = 10.58NALNQLSPEE667 pKa = 4.09IYY669 pKa = 10.52SNAEE673 pKa = 3.12IAALYY678 pKa = 10.62SSLAFSNSLLSCKK691 pKa = 10.5VNGTGTGTAAIIRR704 pKa = 11.84EE705 pKa = 4.68GQCLWAGASANFVDD719 pKa = 4.82TGTTSDD725 pKa = 4.03QIGFTQTTGLFNAGAQVALDD745 pKa = 3.85DD746 pKa = 3.66VWRR749 pKa = 11.84VGFGAGYY756 pKa = 10.01QSSSLQTATNAQSEE770 pKa = 4.73GSLAQAGLSVKK781 pKa = 10.23YY782 pKa = 10.83NPGPLLLAGAVSGGGAWYY800 pKa = 7.87DD801 pKa = 3.63TTRR804 pKa = 11.84TMAFGGFTGVAEE816 pKa = 4.56GDD818 pKa = 3.3QDD820 pKa = 3.78VGIVSGSLRR829 pKa = 11.84AAYY832 pKa = 10.32VLGSPHH838 pKa = 7.45LYY840 pKa = 10.07YY841 pKa = 10.72KK842 pKa = 10.61PILDD846 pKa = 4.5LGLTHH851 pKa = 7.27LEE853 pKa = 3.9LGGFTEE859 pKa = 4.57SGGGGAALTVAGGGQTVFSLAPTLEE884 pKa = 4.4VGSEE888 pKa = 3.26WWMSNGTLIRR898 pKa = 11.84PMIRR902 pKa = 11.84GGAIWYY908 pKa = 9.4DD909 pKa = 3.38GADD912 pKa = 3.68FALTAAFAGAPLGVSPFTINTNIDD936 pKa = 3.26EE937 pKa = 4.35VMGLVGAGVEE947 pKa = 4.46VINGGDD953 pKa = 3.07AALRR957 pKa = 11.84FSYY960 pKa = 10.38DD961 pKa = 3.09GQLGEE966 pKa = 4.3TTQIHH971 pKa = 4.78TVGVKK976 pKa = 10.14GSARR980 pKa = 11.84FF981 pKa = 3.39
Molecular weight: 97.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I3KQD6|A0A6I3KQD6_9RHIZ Delta-aminolevulinic acid dehydratase OS=Hyphomicrobium sp. xq OX=2665159 GN=hemB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.26RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.73GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MQTTGGVRR29 pKa = 11.84VLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.26RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.73GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MQTTGGVRR29 pKa = 11.84VLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1132951 |
29 |
4183 |
309.2 |
33.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.811 ± 0.058 | 0.871 ± 0.018 |
5.633 ± 0.032 | 5.671 ± 0.042 |
3.531 ± 0.022 | 8.57 ± 0.072 |
1.963 ± 0.022 | 4.935 ± 0.029 |
3.774 ± 0.04 | 9.826 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.237 ± 0.021 | 2.652 ± 0.034 |
5.313 ± 0.038 | 3.134 ± 0.024 |
7.031 ± 0.051 | 5.467 ± 0.038 |
5.401 ± 0.04 | 7.563 ± 0.039 |
1.309 ± 0.018 | 2.306 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
