
Desulfuromonas acetoxidans (strain DSM 684 / 11070)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Desulfuromonas; Desulfuromonas acetoxidans
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
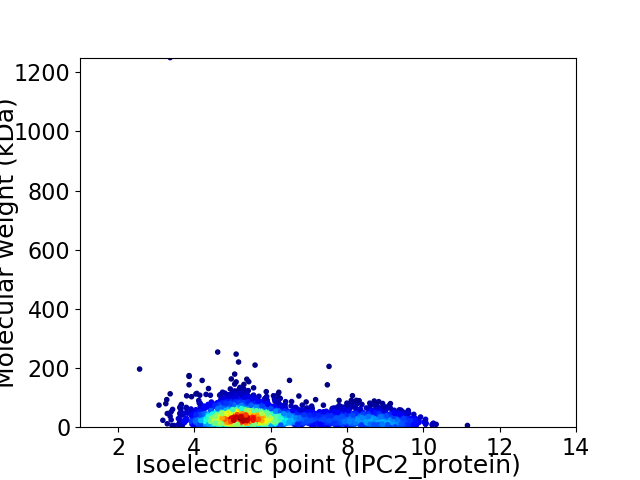
Virtual 2D-PAGE plot for 3204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1K3J6|Q1K3J6_DESA6 Hydroxylamine reductase OS=Desulfuromonas acetoxidans (strain DSM 684 / 11070) OX=281689 GN=hcp PE=3 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84SLKK5 pKa = 9.15MACPTLCVMLLTLCWSGLAAAYY27 pKa = 10.15DD28 pKa = 4.35PIIHH32 pKa = 6.21SEE34 pKa = 4.61FYY36 pKa = 10.47YY37 pKa = 11.12GPDD40 pKa = 3.37SEE42 pKa = 4.72VHH44 pKa = 5.4VVGDD48 pKa = 3.72MLSVPEE54 pKa = 4.75FSLTGEE60 pKa = 4.6GIDD63 pKa = 3.6VVFSGLEE70 pKa = 3.64EE71 pKa = 4.5GGGVEE76 pKa = 4.58KK77 pKa = 11.06YY78 pKa = 10.64AVLLEE83 pKa = 4.11YY84 pKa = 10.79DD85 pKa = 3.97SGSADD90 pKa = 3.5LRR92 pKa = 11.84DD93 pKa = 3.79ATVSLTDD100 pKa = 3.12ATFFFDD106 pKa = 3.51TSGGNEE112 pKa = 4.13DD113 pKa = 3.9FNFAVGMKK121 pKa = 10.41GFDD124 pKa = 3.24EE125 pKa = 4.49SLISLAEE132 pKa = 4.28GSSLSVDD139 pKa = 2.68IASYY143 pKa = 9.72IGYY146 pKa = 9.44GIEE149 pKa = 4.07VLRR152 pKa = 11.84SSATINLDD160 pKa = 3.25GGSTIDD166 pKa = 3.26VSGSVEE172 pKa = 4.24DD173 pKa = 4.49PVTFPDD179 pKa = 3.61SAPSRR184 pKa = 11.84FGYY187 pKa = 10.12GINAGTPGGYY197 pKa = 10.4VNLSLDD203 pKa = 3.47HH204 pKa = 6.75GSAINVEE211 pKa = 4.34TSAYY215 pKa = 10.14AAGDD219 pKa = 3.69TGTSSASSAGARR231 pKa = 11.84LEE233 pKa = 4.21GDD235 pKa = 3.97VVTVALDD242 pKa = 3.42NEE244 pKa = 4.62STIRR248 pKa = 11.84TVATRR253 pKa = 11.84NIASDD258 pKa = 3.62GVTVANARR266 pKa = 11.84NLRR269 pKa = 11.84LGSISNSTIAVAGNSLISAQAFAEE293 pKa = 4.21EE294 pKa = 4.52GAAYY298 pKa = 9.8AYY300 pKa = 10.49VLQTYY305 pKa = 10.42GSGASEE311 pKa = 4.26FLLSDD316 pKa = 3.54STFSATATADD326 pKa = 3.56GYY328 pKa = 11.83ADD330 pKa = 3.59ALGAYY335 pKa = 9.19FSQPEE340 pKa = 3.9VRR342 pKa = 11.84ATLTRR347 pKa = 11.84STIDD351 pKa = 2.72VDD353 pKa = 5.04AISSSYY359 pKa = 10.06GARR362 pKa = 11.84ASGFVGQSNSDD373 pKa = 2.98VRR375 pKa = 11.84LEE377 pKa = 4.08MEE379 pKa = 4.29GATIDD384 pKa = 4.9VLAQTQGDD392 pKa = 3.97FANATGVMIIGATNLDD408 pKa = 3.12IDD410 pKa = 4.25IADD413 pKa = 3.82STVDD417 pKa = 3.38TFAQAAGGEE426 pKa = 4.18AEE428 pKa = 4.47AVGFSLVGSTTISSLVLNRR447 pKa = 11.84SYY449 pKa = 11.07ILSSAVATDD458 pKa = 3.31SDD460 pKa = 4.14ASVGEE465 pKa = 4.37AYY467 pKa = 10.27AAGLIIGVDD476 pKa = 3.74TVAEE480 pKa = 4.4NISLVGSHH488 pKa = 6.4LMSSAHH494 pKa = 5.77AQTGEE499 pKa = 3.9AVSYY503 pKa = 10.6GVFVEE508 pKa = 5.03DD509 pKa = 4.8YY510 pKa = 10.63YY511 pKa = 11.71GGTPEE516 pKa = 3.95VTLNLDD522 pKa = 3.13AHH524 pKa = 6.54SGILADD530 pKa = 3.61WAVYY534 pKa = 8.33TADD537 pKa = 4.55GVVHH541 pKa = 6.73LNNSGTLAGRR551 pKa = 11.84LLVSDD556 pKa = 4.09LTNNVGGRR564 pKa = 11.84FMLLLDD570 pKa = 5.02SDD572 pKa = 4.71DD573 pKa = 4.55LLIGEE578 pKa = 5.12DD579 pKa = 3.56GVGYY583 pKa = 10.73FEE585 pKa = 6.64DD586 pKa = 3.62EE587 pKa = 4.03DD588 pKa = 5.64FYY590 pKa = 11.75FSVADD595 pKa = 4.06RR596 pKa = 11.84AEE598 pKa = 4.25LADD601 pKa = 3.42GTTFLVYY608 pKa = 10.36PSDD611 pKa = 3.48NLGIVSEE618 pKa = 4.52GDD620 pKa = 3.48STSIALLSAGEE631 pKa = 4.15GDD633 pKa = 3.19WTLEE637 pKa = 3.96NLSLEE642 pKa = 4.61TPFDD646 pKa = 3.56TPMIGLEE653 pKa = 4.13LTEE656 pKa = 4.19NAAGNQLIATATFLTPEE673 pKa = 4.03QAGLSANATAAYY685 pKa = 7.53EE686 pKa = 3.99AAVDD690 pKa = 3.96GGLFTPTSSPEE701 pKa = 3.58EE702 pKa = 3.8WSPNVSGAFVVGMAHH717 pKa = 6.25TLMVSNMNIGNRR729 pKa = 11.84LGMLSGMNSGDD740 pKa = 3.98EE741 pKa = 4.04IAASNGMWFSARR753 pKa = 11.84FTDD756 pKa = 4.37AEE758 pKa = 3.84QDD760 pKa = 3.24RR761 pKa = 11.84RR762 pKa = 11.84DD763 pKa = 4.04GIEE766 pKa = 4.8GFDD769 pKa = 4.28ADD771 pKa = 4.0TDD773 pKa = 4.0ALSFGFDD780 pKa = 2.97RR781 pKa = 11.84EE782 pKa = 4.17FGNVVLGVAYY792 pKa = 9.48TRR794 pKa = 11.84GNSDD798 pKa = 3.17AKK800 pKa = 11.36ANDD803 pKa = 3.29ASADD807 pKa = 3.61FEE809 pKa = 4.3MSDD812 pKa = 4.51NLFSLYY818 pKa = 10.72GSYY821 pKa = 10.92DD822 pKa = 2.91GGTWYY827 pKa = 10.85SEE829 pKa = 3.98AVVSVGFGSVDD840 pKa = 3.25SLRR843 pKa = 11.84SVGSDD848 pKa = 4.03DD849 pKa = 3.77YY850 pKa = 12.04AGDD853 pKa = 3.98YY854 pKa = 11.19DD855 pKa = 3.94STSYY859 pKa = 9.57STLVEE864 pKa = 4.38SGCKK868 pKa = 9.38ISAAGWEE875 pKa = 4.21IYY877 pKa = 9.93PYY879 pKa = 9.83LTLEE883 pKa = 4.24YY884 pKa = 10.39SSKK887 pKa = 11.09DD888 pKa = 3.21YY889 pKa = 11.32DD890 pKa = 3.86SYY892 pKa = 11.7TEE894 pKa = 4.17TGGSLALHH902 pKa = 6.09VRR904 pKa = 11.84SKK906 pKa = 10.87DD907 pKa = 3.63YY908 pKa = 11.32EE909 pKa = 4.26VFTAGVGARR918 pKa = 11.84LQKK921 pKa = 10.56DD922 pKa = 4.09FQRR925 pKa = 11.84SWGIVTPEE933 pKa = 3.74VSGQLAYY940 pKa = 10.39DD941 pKa = 3.72VEE943 pKa = 4.32NDD945 pKa = 4.5RR946 pKa = 11.84IVSTANFVGGTTSFVAKK963 pKa = 10.18GIEE966 pKa = 3.9PSEE969 pKa = 4.24TSWDD973 pKa = 3.24LGAALTVASLGEE985 pKa = 4.12QNVSLRR991 pKa = 11.84LGYY994 pKa = 10.49DD995 pKa = 3.12YY996 pKa = 11.49AGRR999 pKa = 11.84QDD1001 pKa = 3.84FAAHH1005 pKa = 6.4SFTGKK1010 pKa = 9.76VRR1012 pKa = 11.84FEE1014 pKa = 4.22FF1015 pKa = 4.5
MM1 pKa = 7.98RR2 pKa = 11.84SLKK5 pKa = 9.15MACPTLCVMLLTLCWSGLAAAYY27 pKa = 10.15DD28 pKa = 4.35PIIHH32 pKa = 6.21SEE34 pKa = 4.61FYY36 pKa = 10.47YY37 pKa = 11.12GPDD40 pKa = 3.37SEE42 pKa = 4.72VHH44 pKa = 5.4VVGDD48 pKa = 3.72MLSVPEE54 pKa = 4.75FSLTGEE60 pKa = 4.6GIDD63 pKa = 3.6VVFSGLEE70 pKa = 3.64EE71 pKa = 4.5GGGVEE76 pKa = 4.58KK77 pKa = 11.06YY78 pKa = 10.64AVLLEE83 pKa = 4.11YY84 pKa = 10.79DD85 pKa = 3.97SGSADD90 pKa = 3.5LRR92 pKa = 11.84DD93 pKa = 3.79ATVSLTDD100 pKa = 3.12ATFFFDD106 pKa = 3.51TSGGNEE112 pKa = 4.13DD113 pKa = 3.9FNFAVGMKK121 pKa = 10.41GFDD124 pKa = 3.24EE125 pKa = 4.49SLISLAEE132 pKa = 4.28GSSLSVDD139 pKa = 2.68IASYY143 pKa = 9.72IGYY146 pKa = 9.44GIEE149 pKa = 4.07VLRR152 pKa = 11.84SSATINLDD160 pKa = 3.25GGSTIDD166 pKa = 3.26VSGSVEE172 pKa = 4.24DD173 pKa = 4.49PVTFPDD179 pKa = 3.61SAPSRR184 pKa = 11.84FGYY187 pKa = 10.12GINAGTPGGYY197 pKa = 10.4VNLSLDD203 pKa = 3.47HH204 pKa = 6.75GSAINVEE211 pKa = 4.34TSAYY215 pKa = 10.14AAGDD219 pKa = 3.69TGTSSASSAGARR231 pKa = 11.84LEE233 pKa = 4.21GDD235 pKa = 3.97VVTVALDD242 pKa = 3.42NEE244 pKa = 4.62STIRR248 pKa = 11.84TVATRR253 pKa = 11.84NIASDD258 pKa = 3.62GVTVANARR266 pKa = 11.84NLRR269 pKa = 11.84LGSISNSTIAVAGNSLISAQAFAEE293 pKa = 4.21EE294 pKa = 4.52GAAYY298 pKa = 9.8AYY300 pKa = 10.49VLQTYY305 pKa = 10.42GSGASEE311 pKa = 4.26FLLSDD316 pKa = 3.54STFSATATADD326 pKa = 3.56GYY328 pKa = 11.83ADD330 pKa = 3.59ALGAYY335 pKa = 9.19FSQPEE340 pKa = 3.9VRR342 pKa = 11.84ATLTRR347 pKa = 11.84STIDD351 pKa = 2.72VDD353 pKa = 5.04AISSSYY359 pKa = 10.06GARR362 pKa = 11.84ASGFVGQSNSDD373 pKa = 2.98VRR375 pKa = 11.84LEE377 pKa = 4.08MEE379 pKa = 4.29GATIDD384 pKa = 4.9VLAQTQGDD392 pKa = 3.97FANATGVMIIGATNLDD408 pKa = 3.12IDD410 pKa = 4.25IADD413 pKa = 3.82STVDD417 pKa = 3.38TFAQAAGGEE426 pKa = 4.18AEE428 pKa = 4.47AVGFSLVGSTTISSLVLNRR447 pKa = 11.84SYY449 pKa = 11.07ILSSAVATDD458 pKa = 3.31SDD460 pKa = 4.14ASVGEE465 pKa = 4.37AYY467 pKa = 10.27AAGLIIGVDD476 pKa = 3.74TVAEE480 pKa = 4.4NISLVGSHH488 pKa = 6.4LMSSAHH494 pKa = 5.77AQTGEE499 pKa = 3.9AVSYY503 pKa = 10.6GVFVEE508 pKa = 5.03DD509 pKa = 4.8YY510 pKa = 10.63YY511 pKa = 11.71GGTPEE516 pKa = 3.95VTLNLDD522 pKa = 3.13AHH524 pKa = 6.54SGILADD530 pKa = 3.61WAVYY534 pKa = 8.33TADD537 pKa = 4.55GVVHH541 pKa = 6.73LNNSGTLAGRR551 pKa = 11.84LLVSDD556 pKa = 4.09LTNNVGGRR564 pKa = 11.84FMLLLDD570 pKa = 5.02SDD572 pKa = 4.71DD573 pKa = 4.55LLIGEE578 pKa = 5.12DD579 pKa = 3.56GVGYY583 pKa = 10.73FEE585 pKa = 6.64DD586 pKa = 3.62EE587 pKa = 4.03DD588 pKa = 5.64FYY590 pKa = 11.75FSVADD595 pKa = 4.06RR596 pKa = 11.84AEE598 pKa = 4.25LADD601 pKa = 3.42GTTFLVYY608 pKa = 10.36PSDD611 pKa = 3.48NLGIVSEE618 pKa = 4.52GDD620 pKa = 3.48STSIALLSAGEE631 pKa = 4.15GDD633 pKa = 3.19WTLEE637 pKa = 3.96NLSLEE642 pKa = 4.61TPFDD646 pKa = 3.56TPMIGLEE653 pKa = 4.13LTEE656 pKa = 4.19NAAGNQLIATATFLTPEE673 pKa = 4.03QAGLSANATAAYY685 pKa = 7.53EE686 pKa = 3.99AAVDD690 pKa = 3.96GGLFTPTSSPEE701 pKa = 3.58EE702 pKa = 3.8WSPNVSGAFVVGMAHH717 pKa = 6.25TLMVSNMNIGNRR729 pKa = 11.84LGMLSGMNSGDD740 pKa = 3.98EE741 pKa = 4.04IAASNGMWFSARR753 pKa = 11.84FTDD756 pKa = 4.37AEE758 pKa = 3.84QDD760 pKa = 3.24RR761 pKa = 11.84RR762 pKa = 11.84DD763 pKa = 4.04GIEE766 pKa = 4.8GFDD769 pKa = 4.28ADD771 pKa = 4.0TDD773 pKa = 4.0ALSFGFDD780 pKa = 2.97RR781 pKa = 11.84EE782 pKa = 4.17FGNVVLGVAYY792 pKa = 9.48TRR794 pKa = 11.84GNSDD798 pKa = 3.17AKK800 pKa = 11.36ANDD803 pKa = 3.29ASADD807 pKa = 3.61FEE809 pKa = 4.3MSDD812 pKa = 4.51NLFSLYY818 pKa = 10.72GSYY821 pKa = 10.92DD822 pKa = 2.91GGTWYY827 pKa = 10.85SEE829 pKa = 3.98AVVSVGFGSVDD840 pKa = 3.25SLRR843 pKa = 11.84SVGSDD848 pKa = 4.03DD849 pKa = 3.77YY850 pKa = 12.04AGDD853 pKa = 3.98YY854 pKa = 11.19DD855 pKa = 3.94STSYY859 pKa = 9.57STLVEE864 pKa = 4.38SGCKK868 pKa = 9.38ISAAGWEE875 pKa = 4.21IYY877 pKa = 9.93PYY879 pKa = 9.83LTLEE883 pKa = 4.24YY884 pKa = 10.39SSKK887 pKa = 11.09DD888 pKa = 3.21YY889 pKa = 11.32DD890 pKa = 3.86SYY892 pKa = 11.7TEE894 pKa = 4.17TGGSLALHH902 pKa = 6.09VRR904 pKa = 11.84SKK906 pKa = 10.87DD907 pKa = 3.63YY908 pKa = 11.32EE909 pKa = 4.26VFTAGVGARR918 pKa = 11.84LQKK921 pKa = 10.56DD922 pKa = 4.09FQRR925 pKa = 11.84SWGIVTPEE933 pKa = 3.74VSGQLAYY940 pKa = 10.39DD941 pKa = 3.72VEE943 pKa = 4.32NDD945 pKa = 4.5RR946 pKa = 11.84IVSTANFVGGTTSFVAKK963 pKa = 10.18GIEE966 pKa = 3.9PSEE969 pKa = 4.24TSWDD973 pKa = 3.24LGAALTVASLGEE985 pKa = 4.12QNVSLRR991 pKa = 11.84LGYY994 pKa = 10.49DD995 pKa = 3.12YY996 pKa = 11.49AGRR999 pKa = 11.84QDD1001 pKa = 3.84FAAHH1005 pKa = 6.4SFTGKK1010 pKa = 9.76VRR1012 pKa = 11.84FEE1014 pKa = 4.22FF1015 pKa = 4.5
Molecular weight: 106.0 kDa
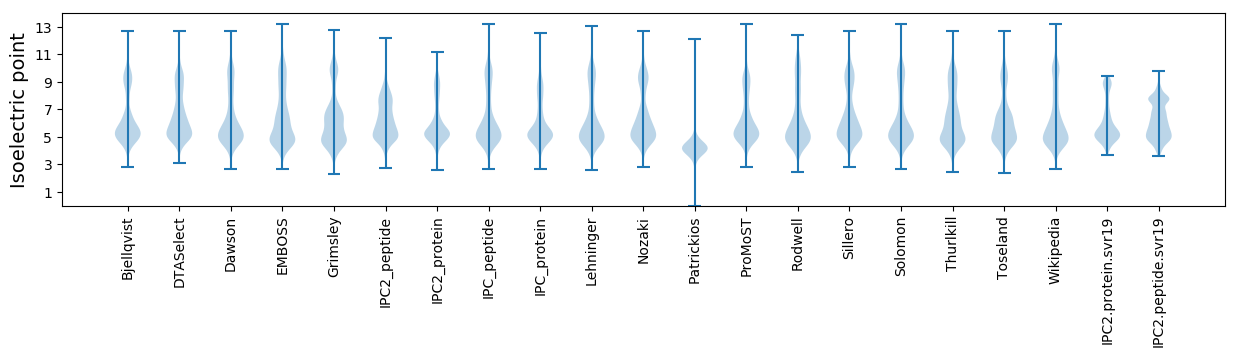
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1JZH4|Q1JZH4_DESA6 7-cyano-7-deazaguanine synthase OS=Desulfuromonas acetoxidans (strain DSM 684 / 11070) OX=281689 GN=queC PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.19QPSRR9 pKa = 11.84VSRR12 pKa = 11.84KK13 pKa = 7.52RR14 pKa = 11.84THH16 pKa = 6.3GFRR19 pKa = 11.84KK20 pKa = 9.97RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84NVIKK32 pKa = 10.48RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.71VLAATISSKK49 pKa = 11.21
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.19QPSRR9 pKa = 11.84VSRR12 pKa = 11.84KK13 pKa = 7.52RR14 pKa = 11.84THH16 pKa = 6.3GFRR19 pKa = 11.84KK20 pKa = 9.97RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84NVIKK32 pKa = 10.48RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.71VLAATISSKK49 pKa = 11.21
Molecular weight: 5.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103131 |
36 |
12388 |
344.3 |
38.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.798 ± 0.055 | 1.378 ± 0.024 |
5.949 ± 0.068 | 6.333 ± 0.048 |
3.907 ± 0.039 | 7.18 ± 0.073 |
2.391 ± 0.031 | 5.819 ± 0.035 |
4.242 ± 0.062 | 10.766 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.029 | 3.448 ± 0.029 |
4.129 ± 0.049 | 4.693 ± 0.045 |
5.637 ± 0.067 | 6.049 ± 0.066 |
5.472 ± 0.07 | 7.352 ± 0.051 |
1.108 ± 0.02 | 2.859 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
