
Woeseia oceani
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Woeseiaceae; Woeseia
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
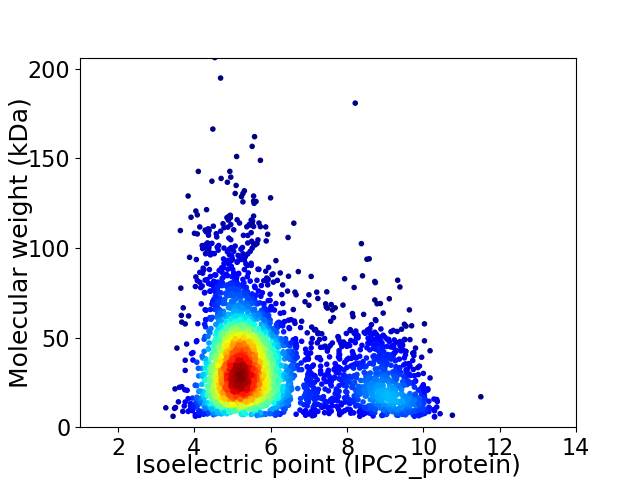
Virtual 2D-PAGE plot for 3565 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A193LBU3|A0A193LBU3_9GAMM Transcriptional regulator OS=Woeseia oceani OX=1548547 GN=BA177_01085 PE=4 SV=1
MM1 pKa = 7.05RR2 pKa = 11.84TKK4 pKa = 10.66LALTAVVASIVAAGCGSGDD23 pKa = 3.52INIDD27 pKa = 3.26PVTSVTNSNNTTNTGGNSGTTNPCASYY54 pKa = 11.12QNTGGQSIQGTYY66 pKa = 10.88DD67 pKa = 3.36GTHH70 pKa = 5.87CTYY73 pKa = 10.97SPSFVDD79 pKa = 3.44AGNNLMVDD87 pKa = 4.26LTLADD92 pKa = 4.27LPNDD96 pKa = 3.93GAHH99 pKa = 6.43LFEE102 pKa = 5.68GSLFVGEE109 pKa = 4.51SHH111 pKa = 7.89DD112 pKa = 4.44NDD114 pKa = 3.99ADD116 pKa = 3.85LAAAGIAEE124 pKa = 4.7GGDD127 pKa = 3.81GPVLTVEE134 pKa = 4.77AGATLALQSSASFMIINRR152 pKa = 11.84GSQLFAVGRR161 pKa = 11.84ADD163 pKa = 3.99APITFTSLSDD173 pKa = 3.24INGTVGPEE181 pKa = 4.69DD182 pKa = 3.58VQQWGGMVINGFGITNKK199 pKa = 9.87CAYY202 pKa = 8.0TGSVATNDD210 pKa = 3.88LATSDD215 pKa = 3.93CHH217 pKa = 8.07VDD219 pKa = 3.23AEE221 pKa = 4.75GAAGLDD227 pKa = 3.53EE228 pKa = 4.32SQYY231 pKa = 11.76GGDD234 pKa = 4.07NNDD237 pKa = 3.79DD238 pKa = 3.31SSGRR242 pKa = 11.84MEE244 pKa = 4.38YY245 pKa = 10.83VVVKK249 pKa = 8.84HH250 pKa = 6.12TGATVGNGDD259 pKa = 4.29EE260 pKa = 4.65LNGITFGGVGRR271 pKa = 11.84NTIVNNLQVYY281 pKa = 7.69STFDD285 pKa = 3.23DD286 pKa = 5.04GIEE289 pKa = 4.08MFGGAVNFNNFVGMYY304 pKa = 9.88VRR306 pKa = 11.84DD307 pKa = 3.76DD308 pKa = 5.08AIDD311 pKa = 3.42IDD313 pKa = 4.06EE314 pKa = 5.49GYY316 pKa = 10.82SGTITNALVIQSEE329 pKa = 4.59TDD331 pKa = 3.22GNHH334 pKa = 6.8CIEE337 pKa = 4.76ADD339 pKa = 4.17GIGGFDD345 pKa = 4.11DD346 pKa = 4.97LLPAEE351 pKa = 4.46VEE353 pKa = 4.1AMISQGINSRR363 pKa = 11.84FVINNLTCIISPNGAATATHH383 pKa = 7.11DD384 pKa = 4.31PGAGWRR390 pKa = 11.84LRR392 pKa = 11.84EE393 pKa = 4.02GLFAEE398 pKa = 4.93INDD401 pKa = 3.93SLLIGSMPANDD412 pKa = 3.04QVSSNDD418 pKa = 3.55NYY420 pKa = 10.93CLRR423 pKa = 11.84IDD425 pKa = 4.77NRR427 pKa = 11.84TQQAALDD434 pKa = 4.04GDD436 pKa = 3.97LTLNSVVFACAEE448 pKa = 3.92ATRR451 pKa = 11.84GQTFADD457 pKa = 3.93GTTTEE462 pKa = 4.22RR463 pKa = 11.84SFAEE467 pKa = 3.74AGGNIFATIANNTSVDD483 pKa = 3.69PTAMNDD489 pKa = 3.07AGLQLLEE496 pKa = 4.34GTQPIYY502 pKa = 10.93SIGYY506 pKa = 5.37TTMQVDD512 pKa = 4.73GAAPAGAPQSGTFIGGLSLSEE533 pKa = 4.38TDD535 pKa = 2.97WTMGWTFGLHH545 pKa = 6.46PDD547 pKa = 3.83NRR549 pKa = 11.84SQALWFEE556 pKa = 4.42
MM1 pKa = 7.05RR2 pKa = 11.84TKK4 pKa = 10.66LALTAVVASIVAAGCGSGDD23 pKa = 3.52INIDD27 pKa = 3.26PVTSVTNSNNTTNTGGNSGTTNPCASYY54 pKa = 11.12QNTGGQSIQGTYY66 pKa = 10.88DD67 pKa = 3.36GTHH70 pKa = 5.87CTYY73 pKa = 10.97SPSFVDD79 pKa = 3.44AGNNLMVDD87 pKa = 4.26LTLADD92 pKa = 4.27LPNDD96 pKa = 3.93GAHH99 pKa = 6.43LFEE102 pKa = 5.68GSLFVGEE109 pKa = 4.51SHH111 pKa = 7.89DD112 pKa = 4.44NDD114 pKa = 3.99ADD116 pKa = 3.85LAAAGIAEE124 pKa = 4.7GGDD127 pKa = 3.81GPVLTVEE134 pKa = 4.77AGATLALQSSASFMIINRR152 pKa = 11.84GSQLFAVGRR161 pKa = 11.84ADD163 pKa = 3.99APITFTSLSDD173 pKa = 3.24INGTVGPEE181 pKa = 4.69DD182 pKa = 3.58VQQWGGMVINGFGITNKK199 pKa = 9.87CAYY202 pKa = 8.0TGSVATNDD210 pKa = 3.88LATSDD215 pKa = 3.93CHH217 pKa = 8.07VDD219 pKa = 3.23AEE221 pKa = 4.75GAAGLDD227 pKa = 3.53EE228 pKa = 4.32SQYY231 pKa = 11.76GGDD234 pKa = 4.07NNDD237 pKa = 3.79DD238 pKa = 3.31SSGRR242 pKa = 11.84MEE244 pKa = 4.38YY245 pKa = 10.83VVVKK249 pKa = 8.84HH250 pKa = 6.12TGATVGNGDD259 pKa = 4.29EE260 pKa = 4.65LNGITFGGVGRR271 pKa = 11.84NTIVNNLQVYY281 pKa = 7.69STFDD285 pKa = 3.23DD286 pKa = 5.04GIEE289 pKa = 4.08MFGGAVNFNNFVGMYY304 pKa = 9.88VRR306 pKa = 11.84DD307 pKa = 3.76DD308 pKa = 5.08AIDD311 pKa = 3.42IDD313 pKa = 4.06EE314 pKa = 5.49GYY316 pKa = 10.82SGTITNALVIQSEE329 pKa = 4.59TDD331 pKa = 3.22GNHH334 pKa = 6.8CIEE337 pKa = 4.76ADD339 pKa = 4.17GIGGFDD345 pKa = 4.11DD346 pKa = 4.97LLPAEE351 pKa = 4.46VEE353 pKa = 4.1AMISQGINSRR363 pKa = 11.84FVINNLTCIISPNGAATATHH383 pKa = 7.11DD384 pKa = 4.31PGAGWRR390 pKa = 11.84LRR392 pKa = 11.84EE393 pKa = 4.02GLFAEE398 pKa = 4.93INDD401 pKa = 3.93SLLIGSMPANDD412 pKa = 3.04QVSSNDD418 pKa = 3.55NYY420 pKa = 10.93CLRR423 pKa = 11.84IDD425 pKa = 4.77NRR427 pKa = 11.84TQQAALDD434 pKa = 4.04GDD436 pKa = 3.97LTLNSVVFACAEE448 pKa = 3.92ATRR451 pKa = 11.84GQTFADD457 pKa = 3.93GTTTEE462 pKa = 4.22RR463 pKa = 11.84SFAEE467 pKa = 3.74AGGNIFATIANNTSVDD483 pKa = 3.69PTAMNDD489 pKa = 3.07AGLQLLEE496 pKa = 4.34GTQPIYY502 pKa = 10.93SIGYY506 pKa = 5.37TTMQVDD512 pKa = 4.73GAAPAGAPQSGTFIGGLSLSEE533 pKa = 4.38TDD535 pKa = 2.97WTMGWTFGLHH545 pKa = 6.46PDD547 pKa = 3.83NRR549 pKa = 11.84SQALWFEE556 pKa = 4.42
Molecular weight: 57.63 kDa
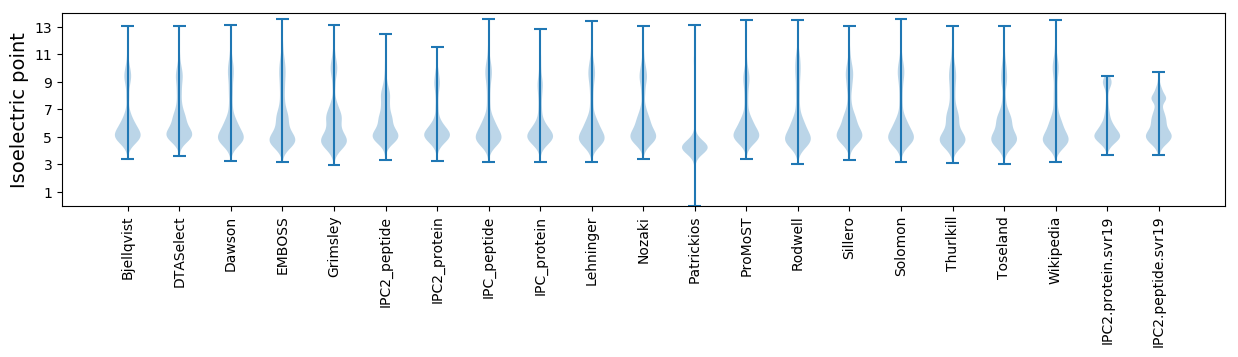
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A193LLG2|A0A193LLG2_9GAMM UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2 6-diaminopimelate ligase OS=Woeseia oceani OX=1548547 GN=murE PE=3 SV=1
MM1 pKa = 7.12ATKK4 pKa = 10.22KK5 pKa = 10.4KK6 pKa = 10.33SSKK9 pKa = 8.96KK10 pKa = 7.15TSKK13 pKa = 10.58RR14 pKa = 11.84KK15 pKa = 7.36TATKK19 pKa = 10.38RR20 pKa = 11.84KK21 pKa = 6.26VTKK24 pKa = 10.48KK25 pKa = 10.01KK26 pKa = 8.2VAKK29 pKa = 10.22RR30 pKa = 11.84KK31 pKa = 8.09VAKK34 pKa = 10.4KK35 pKa = 9.96KK36 pKa = 8.15VAKK39 pKa = 10.35RR40 pKa = 11.84KK41 pKa = 5.75TAKK44 pKa = 10.38KK45 pKa = 9.83KK46 pKa = 8.04VAKK49 pKa = 10.36RR50 pKa = 11.84KK51 pKa = 7.12TKK53 pKa = 10.3KK54 pKa = 9.6KK55 pKa = 10.07AKK57 pKa = 10.03KK58 pKa = 9.6KK59 pKa = 7.94VAKK62 pKa = 10.16RR63 pKa = 11.84KK64 pKa = 7.15TKK66 pKa = 10.59KK67 pKa = 10.07KK68 pKa = 7.92VAKK71 pKa = 10.28RR72 pKa = 11.84KK73 pKa = 7.12TKK75 pKa = 10.45KK76 pKa = 9.7KK77 pKa = 10.16AKK79 pKa = 9.78KK80 pKa = 8.96KK81 pKa = 9.84AKK83 pKa = 10.16KK84 pKa = 9.76KK85 pKa = 7.97VAKK88 pKa = 10.25RR89 pKa = 11.84KK90 pKa = 7.28TKK92 pKa = 10.6KK93 pKa = 8.87KK94 pKa = 8.18TKK96 pKa = 10.19KK97 pKa = 9.75KK98 pKa = 9.46AAKK101 pKa = 9.99KK102 pKa = 8.92KK103 pKa = 6.97TAKK106 pKa = 10.28KK107 pKa = 9.56KK108 pKa = 10.27AKK110 pKa = 9.88KK111 pKa = 9.32KK112 pKa = 9.89AKK114 pKa = 10.21KK115 pKa = 9.86KK116 pKa = 8.69VAKK119 pKa = 9.99KK120 pKa = 8.82KK121 pKa = 10.54AKK123 pKa = 10.25RR124 pKa = 11.84KK125 pKa = 8.65VKK127 pKa = 10.49KK128 pKa = 10.01KK129 pKa = 10.11ASKK132 pKa = 10.4KK133 pKa = 8.89KK134 pKa = 9.15ATRR137 pKa = 11.84KK138 pKa = 6.94TRR140 pKa = 11.84KK141 pKa = 9.32KK142 pKa = 10.45ASKK145 pKa = 10.29KK146 pKa = 9.93KK147 pKa = 10.32
MM1 pKa = 7.12ATKK4 pKa = 10.22KK5 pKa = 10.4KK6 pKa = 10.33SSKK9 pKa = 8.96KK10 pKa = 7.15TSKK13 pKa = 10.58RR14 pKa = 11.84KK15 pKa = 7.36TATKK19 pKa = 10.38RR20 pKa = 11.84KK21 pKa = 6.26VTKK24 pKa = 10.48KK25 pKa = 10.01KK26 pKa = 8.2VAKK29 pKa = 10.22RR30 pKa = 11.84KK31 pKa = 8.09VAKK34 pKa = 10.4KK35 pKa = 9.96KK36 pKa = 8.15VAKK39 pKa = 10.35RR40 pKa = 11.84KK41 pKa = 5.75TAKK44 pKa = 10.38KK45 pKa = 9.83KK46 pKa = 8.04VAKK49 pKa = 10.36RR50 pKa = 11.84KK51 pKa = 7.12TKK53 pKa = 10.3KK54 pKa = 9.6KK55 pKa = 10.07AKK57 pKa = 10.03KK58 pKa = 9.6KK59 pKa = 7.94VAKK62 pKa = 10.16RR63 pKa = 11.84KK64 pKa = 7.15TKK66 pKa = 10.59KK67 pKa = 10.07KK68 pKa = 7.92VAKK71 pKa = 10.28RR72 pKa = 11.84KK73 pKa = 7.12TKK75 pKa = 10.45KK76 pKa = 9.7KK77 pKa = 10.16AKK79 pKa = 9.78KK80 pKa = 8.96KK81 pKa = 9.84AKK83 pKa = 10.16KK84 pKa = 9.76KK85 pKa = 7.97VAKK88 pKa = 10.25RR89 pKa = 11.84KK90 pKa = 7.28TKK92 pKa = 10.6KK93 pKa = 8.87KK94 pKa = 8.18TKK96 pKa = 10.19KK97 pKa = 9.75KK98 pKa = 9.46AAKK101 pKa = 9.99KK102 pKa = 8.92KK103 pKa = 6.97TAKK106 pKa = 10.28KK107 pKa = 9.56KK108 pKa = 10.27AKK110 pKa = 9.88KK111 pKa = 9.32KK112 pKa = 9.89AKK114 pKa = 10.21KK115 pKa = 9.86KK116 pKa = 8.69VAKK119 pKa = 9.99KK120 pKa = 8.82KK121 pKa = 10.54AKK123 pKa = 10.25RR124 pKa = 11.84KK125 pKa = 8.65VKK127 pKa = 10.49KK128 pKa = 10.01KK129 pKa = 10.11ASKK132 pKa = 10.4KK133 pKa = 8.89KK134 pKa = 9.15ATRR137 pKa = 11.84KK138 pKa = 6.94TRR140 pKa = 11.84KK141 pKa = 9.32KK142 pKa = 10.45ASKK145 pKa = 10.29KK146 pKa = 9.93KK147 pKa = 10.32
Molecular weight: 16.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1201765 |
51 |
1901 |
337.1 |
36.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.096 ± 0.045 | 0.916 ± 0.016 |
6.191 ± 0.032 | 5.933 ± 0.034 |
3.704 ± 0.026 | 7.942 ± 0.034 |
2.032 ± 0.019 | 5.198 ± 0.032 |
3.024 ± 0.033 | 10.319 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.342 ± 0.019 | 3.332 ± 0.024 |
4.563 ± 0.024 | 3.675 ± 0.025 |
6.87 ± 0.03 | 6.169 ± 0.03 |
5.252 ± 0.031 | 7.387 ± 0.032 |
1.331 ± 0.017 | 2.725 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
