
Heterobasidion partitivirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
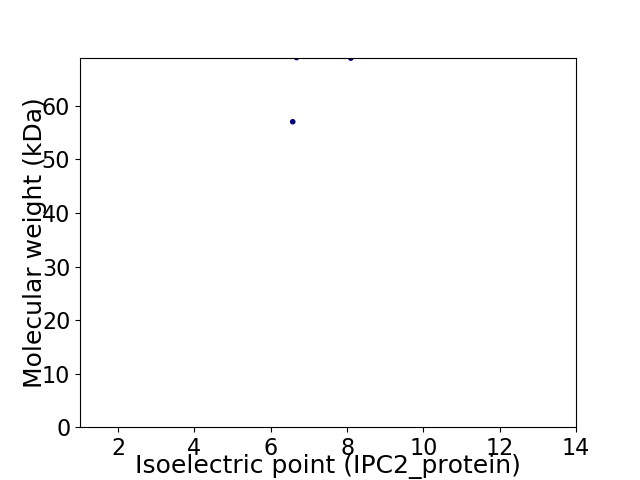
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
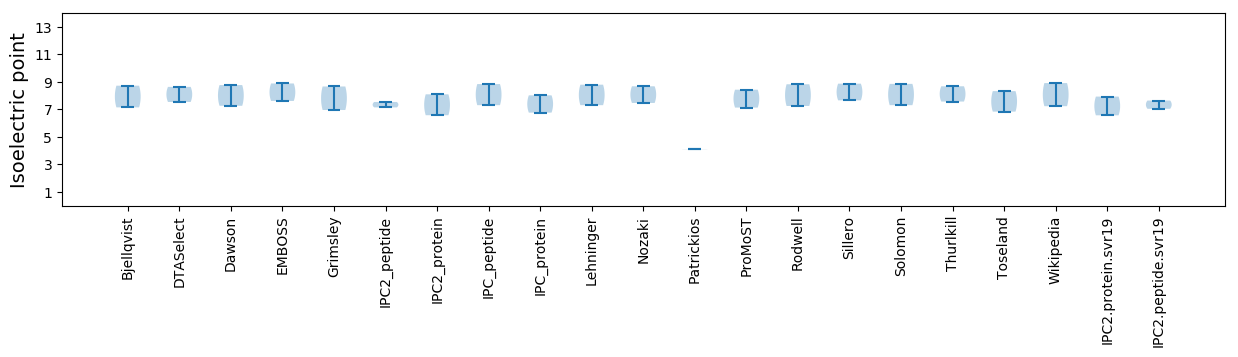
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C1KDH7|C1KDH7_9VIRU Putative coat protein OS=Heterobasidion partitivirus 3 OX=631431 PE=4 SV=1
MM1 pKa = 7.8SSIPDD6 pKa = 3.65FASMNEE12 pKa = 3.94SDD14 pKa = 3.17RR15 pKa = 11.84LKK17 pKa = 11.02YY18 pKa = 10.55FEE20 pKa = 4.5EE21 pKa = 4.04QQKK24 pKa = 9.55KK25 pKa = 7.92QKK27 pKa = 10.51EE28 pKa = 4.25LIDD31 pKa = 3.55QSIKK35 pKa = 10.82SKK37 pKa = 8.81QTMSAMRR44 pKa = 11.84VRR46 pKa = 11.84PVFQQARR53 pKa = 11.84NNVTSVNTSSSVPTKK68 pKa = 10.45AVPPQPFSGTSSSANNMLRR87 pKa = 11.84LAFGEE92 pKa = 4.12NPFRR96 pKa = 11.84PKK98 pKa = 10.41QRR100 pKa = 11.84FGQNGFVPDD109 pKa = 4.01SQHH112 pKa = 6.78LFALLSVMDD121 pKa = 4.77AKK123 pKa = 10.3MASTRR128 pKa = 11.84KK129 pKa = 9.17FVEE132 pKa = 4.78GCEE135 pKa = 3.8FWTPLVSQYY144 pKa = 10.91YY145 pKa = 9.98LSVLIFIQVARR156 pKa = 11.84AKK158 pKa = 10.55HH159 pKa = 4.69EE160 pKa = 4.57AGLLSGEE167 pKa = 4.23LADD170 pKa = 5.78VYY172 pKa = 11.59DD173 pKa = 4.85LFCGHH178 pKa = 6.37SPAINLNALPVPGPFLNMLSQVAAHH203 pKa = 6.81IPHH206 pKa = 6.91LVDD209 pKa = 3.99MDD211 pKa = 4.79NICPIVPNNLEE222 pKa = 4.07ATNTSYY228 pKa = 11.68YY229 pKa = 10.41LYY231 pKa = 10.32TGQCQNLRR239 pKa = 11.84GRR241 pKa = 11.84LPNVPYY247 pKa = 10.47ILDD250 pKa = 3.48QLHH253 pKa = 5.58QLGVLLTANPVGIDD267 pKa = 3.21VANQGRR273 pKa = 11.84VLHH276 pKa = 6.46HH277 pKa = 6.78SFLGVPMFDD286 pKa = 2.93ATVAPGNAAQWNSRR300 pKa = 11.84VAFDD304 pKa = 4.79AGFTASGSFIAVDD317 pKa = 3.61PAARR321 pKa = 11.84HH322 pKa = 5.48PFFLNRR328 pKa = 11.84GLLSQITDD336 pKa = 3.41YY337 pKa = 11.53AALINVTRR345 pKa = 11.84PARR348 pKa = 11.84QNAAAVTPSWTQFLGLDD365 pKa = 3.44NMPFFLQVVRR375 pKa = 11.84IMSWYY380 pKa = 9.16SKK382 pKa = 9.36FWQGSSDD389 pKa = 3.6LMSFTPSGHH398 pKa = 6.39TGGQNTFEE406 pKa = 4.6LVNPPASLNAMTAGGRR422 pKa = 11.84FYY424 pKa = 10.85TLDD427 pKa = 4.2LKK429 pKa = 11.42SKK431 pKa = 9.95GYY433 pKa = 10.16SRR435 pKa = 11.84NPLAPDD441 pKa = 4.14ADD443 pKa = 4.23CFDD446 pKa = 4.39AALGPINLSWTPADD460 pKa = 3.51MFAGKK465 pKa = 9.77LARR468 pKa = 11.84NGAIADD474 pKa = 3.88EE475 pKa = 4.6TTLSNTRR482 pKa = 11.84TAGPYY487 pKa = 9.48FSEE490 pKa = 4.69TIVDD494 pKa = 3.29ITGEE498 pKa = 4.17INPSSAYY505 pKa = 10.34AGILNQYY512 pKa = 8.75YY513 pKa = 9.59YY514 pKa = 11.47SSVALKK520 pKa = 10.59NN521 pKa = 3.48
MM1 pKa = 7.8SSIPDD6 pKa = 3.65FASMNEE12 pKa = 3.94SDD14 pKa = 3.17RR15 pKa = 11.84LKK17 pKa = 11.02YY18 pKa = 10.55FEE20 pKa = 4.5EE21 pKa = 4.04QQKK24 pKa = 9.55KK25 pKa = 7.92QKK27 pKa = 10.51EE28 pKa = 4.25LIDD31 pKa = 3.55QSIKK35 pKa = 10.82SKK37 pKa = 8.81QTMSAMRR44 pKa = 11.84VRR46 pKa = 11.84PVFQQARR53 pKa = 11.84NNVTSVNTSSSVPTKK68 pKa = 10.45AVPPQPFSGTSSSANNMLRR87 pKa = 11.84LAFGEE92 pKa = 4.12NPFRR96 pKa = 11.84PKK98 pKa = 10.41QRR100 pKa = 11.84FGQNGFVPDD109 pKa = 4.01SQHH112 pKa = 6.78LFALLSVMDD121 pKa = 4.77AKK123 pKa = 10.3MASTRR128 pKa = 11.84KK129 pKa = 9.17FVEE132 pKa = 4.78GCEE135 pKa = 3.8FWTPLVSQYY144 pKa = 10.91YY145 pKa = 9.98LSVLIFIQVARR156 pKa = 11.84AKK158 pKa = 10.55HH159 pKa = 4.69EE160 pKa = 4.57AGLLSGEE167 pKa = 4.23LADD170 pKa = 5.78VYY172 pKa = 11.59DD173 pKa = 4.85LFCGHH178 pKa = 6.37SPAINLNALPVPGPFLNMLSQVAAHH203 pKa = 6.81IPHH206 pKa = 6.91LVDD209 pKa = 3.99MDD211 pKa = 4.79NICPIVPNNLEE222 pKa = 4.07ATNTSYY228 pKa = 11.68YY229 pKa = 10.41LYY231 pKa = 10.32TGQCQNLRR239 pKa = 11.84GRR241 pKa = 11.84LPNVPYY247 pKa = 10.47ILDD250 pKa = 3.48QLHH253 pKa = 5.58QLGVLLTANPVGIDD267 pKa = 3.21VANQGRR273 pKa = 11.84VLHH276 pKa = 6.46HH277 pKa = 6.78SFLGVPMFDD286 pKa = 2.93ATVAPGNAAQWNSRR300 pKa = 11.84VAFDD304 pKa = 4.79AGFTASGSFIAVDD317 pKa = 3.61PAARR321 pKa = 11.84HH322 pKa = 5.48PFFLNRR328 pKa = 11.84GLLSQITDD336 pKa = 3.41YY337 pKa = 11.53AALINVTRR345 pKa = 11.84PARR348 pKa = 11.84QNAAAVTPSWTQFLGLDD365 pKa = 3.44NMPFFLQVVRR375 pKa = 11.84IMSWYY380 pKa = 9.16SKK382 pKa = 9.36FWQGSSDD389 pKa = 3.6LMSFTPSGHH398 pKa = 6.39TGGQNTFEE406 pKa = 4.6LVNPPASLNAMTAGGRR422 pKa = 11.84FYY424 pKa = 10.85TLDD427 pKa = 4.2LKK429 pKa = 11.42SKK431 pKa = 9.95GYY433 pKa = 10.16SRR435 pKa = 11.84NPLAPDD441 pKa = 4.14ADD443 pKa = 4.23CFDD446 pKa = 4.39AALGPINLSWTPADD460 pKa = 3.51MFAGKK465 pKa = 9.77LARR468 pKa = 11.84NGAIADD474 pKa = 3.88EE475 pKa = 4.6TTLSNTRR482 pKa = 11.84TAGPYY487 pKa = 9.48FSEE490 pKa = 4.69TIVDD494 pKa = 3.29ITGEE498 pKa = 4.17INPSSAYY505 pKa = 10.34AGILNQYY512 pKa = 8.75YY513 pKa = 9.59YY514 pKa = 11.47SSVALKK520 pKa = 10.59NN521 pKa = 3.48
Molecular weight: 57.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1KDH7|C1KDH7_9VIRU Putative coat protein OS=Heterobasidion partitivirus 3 OX=631431 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.14IFSTLYY8 pKa = 10.31SSFASFAKK16 pKa = 8.47WTGLTDD22 pKa = 3.21PHH24 pKa = 8.0GFEE27 pKa = 4.62HH28 pKa = 6.69NFYY31 pKa = 9.6FTGYY35 pKa = 9.61ADD37 pKa = 3.78KK38 pKa = 10.63KK39 pKa = 10.02IKK41 pKa = 10.7VSINPRR47 pKa = 11.84FQEE50 pKa = 4.23VYY52 pKa = 10.76DD53 pKa = 4.76DD54 pKa = 3.76YY55 pKa = 11.66QSYY58 pKa = 10.33VGKK61 pKa = 10.41FIDD64 pKa = 3.37KK65 pKa = 10.25HH66 pKa = 7.59LPGHH70 pKa = 5.92MAQKK74 pKa = 10.32IKK76 pKa = 10.51FGYY79 pKa = 7.95HH80 pKa = 5.87HH81 pKa = 7.42PVASLPFMITNLKK94 pKa = 10.55KK95 pKa = 10.7GDD97 pKa = 4.16LPDD100 pKa = 4.13HH101 pKa = 6.57PVPHH105 pKa = 6.86DD106 pKa = 3.4QHH108 pKa = 5.34YY109 pKa = 7.47TAARR113 pKa = 11.84KK114 pKa = 8.69AAADD118 pKa = 3.4AFRR121 pKa = 11.84PPRR124 pKa = 11.84LVRR127 pKa = 11.84PVHH130 pKa = 5.99FADD133 pKa = 3.66LRR135 pKa = 11.84YY136 pKa = 10.32YY137 pKa = 9.43KK138 pKa = 10.13WNWHH142 pKa = 6.31PNVEE146 pKa = 4.18EE147 pKa = 4.93PYY149 pKa = 10.86YY150 pKa = 10.76SDD152 pKa = 4.85PKK154 pKa = 8.47LQQYY158 pKa = 9.07VEE160 pKa = 3.99HH161 pKa = 7.34CYY163 pKa = 11.07ALGLIDD169 pKa = 5.33DD170 pKa = 4.7ARR172 pKa = 11.84LSFGNLKK179 pKa = 10.76DD180 pKa = 3.91FVFMDD185 pKa = 3.51TRR187 pKa = 11.84HH188 pKa = 5.57YY189 pKa = 11.06LHH191 pKa = 7.36LIKK194 pKa = 10.87NGSITDD200 pKa = 3.89NNQLWPIMKK209 pKa = 9.55IHH211 pKa = 6.55VKK213 pKa = 9.79PALTEE218 pKa = 3.8PTEE221 pKa = 4.01TKK223 pKa = 9.93IRR225 pKa = 11.84VIYY228 pKa = 9.95GVSKK232 pKa = 10.58RR233 pKa = 11.84HH234 pKa = 6.15ILAQAMFFWPLFRR247 pKa = 11.84YY248 pKa = 9.91YY249 pKa = 9.75IEE251 pKa = 4.36EE252 pKa = 4.16HH253 pKa = 6.1TSPLLWGNEE262 pKa = 3.87TFTGGMLKK270 pKa = 9.36IHH272 pKa = 6.27NLISVPRR279 pKa = 11.84LYY281 pKa = 10.81SQTYY285 pKa = 8.02LTVDD289 pKa = 3.2WSGFDD294 pKa = 3.54LRR296 pKa = 11.84SLFTIQRR303 pKa = 11.84EE304 pKa = 4.15IFDD307 pKa = 3.37DD308 pKa = 2.94WRR310 pKa = 11.84TYY312 pKa = 11.32FDD314 pKa = 3.47FTAYY318 pKa = 9.66IPTRR322 pKa = 11.84TYY324 pKa = 10.71PDD326 pKa = 3.51SKK328 pKa = 9.65TDD330 pKa = 4.41PIRR333 pKa = 11.84MEE335 pKa = 5.12RR336 pKa = 11.84LWNWQRR342 pKa = 11.84DD343 pKa = 3.86ACFKK347 pKa = 10.6MPFVLPDD354 pKa = 3.04RR355 pKa = 11.84TTYY358 pKa = 11.13ARR360 pKa = 11.84LFRR363 pKa = 11.84SIPSGLFVTQFLDD376 pKa = 3.27SHH378 pKa = 6.99YY379 pKa = 9.9NLIMIYY385 pKa = 9.88TILSAMGFDD394 pKa = 3.64ITNLMILVQGDD405 pKa = 3.69DD406 pKa = 3.75SLIHH410 pKa = 6.62LKK412 pKa = 10.59FFLPADD418 pKa = 3.26QHH420 pKa = 6.72DD421 pKa = 4.03AFKK424 pKa = 11.49AEE426 pKa = 4.15FEE428 pKa = 4.29RR429 pKa = 11.84LAKK432 pKa = 10.7YY433 pKa = 10.63YY434 pKa = 9.68FDD436 pKa = 5.56HH437 pKa = 7.21IARR440 pKa = 11.84PEE442 pKa = 4.02KK443 pKa = 9.8THH445 pKa = 5.29VTNSPNEE452 pKa = 4.07VEE454 pKa = 4.28VLGYY458 pKa = 9.19TNNNGYY464 pKa = 9.58PSRR467 pKa = 11.84DD468 pKa = 3.2MTKK471 pKa = 10.55LVAQLYY477 pKa = 9.88HH478 pKa = 6.62PRR480 pKa = 11.84NVDD483 pKa = 3.01KK484 pKa = 10.98TSWKK488 pKa = 9.47SLLMAKK494 pKa = 9.82VCGFAYY500 pKa = 10.29ASCYY504 pKa = 9.75QDD506 pKa = 3.06SQVIDD511 pKa = 4.18LLRR514 pKa = 11.84SIYY517 pKa = 10.99NNLASKK523 pKa = 10.39GFKK526 pKa = 9.49PKK528 pKa = 9.97SGRR531 pKa = 11.84VMRR534 pKa = 11.84DD535 pKa = 2.66IILFGEE541 pKa = 4.59SEE543 pKa = 4.55FEE545 pKa = 4.17VPTDD549 pKa = 4.18HH550 pKa = 7.52FPTLNDD556 pKa = 2.85VTKK559 pKa = 10.09YY560 pKa = 9.69FRR562 pKa = 11.84RR563 pKa = 11.84PYY565 pKa = 10.82VRR567 pKa = 11.84TQRR570 pKa = 11.84DD571 pKa = 3.12ADD573 pKa = 3.77SYY575 pKa = 10.3FPSWHH580 pKa = 6.42FNDD583 pKa = 3.7VFF585 pKa = 4.39
MM1 pKa = 7.69KK2 pKa = 10.14IFSTLYY8 pKa = 10.31SSFASFAKK16 pKa = 8.47WTGLTDD22 pKa = 3.21PHH24 pKa = 8.0GFEE27 pKa = 4.62HH28 pKa = 6.69NFYY31 pKa = 9.6FTGYY35 pKa = 9.61ADD37 pKa = 3.78KK38 pKa = 10.63KK39 pKa = 10.02IKK41 pKa = 10.7VSINPRR47 pKa = 11.84FQEE50 pKa = 4.23VYY52 pKa = 10.76DD53 pKa = 4.76DD54 pKa = 3.76YY55 pKa = 11.66QSYY58 pKa = 10.33VGKK61 pKa = 10.41FIDD64 pKa = 3.37KK65 pKa = 10.25HH66 pKa = 7.59LPGHH70 pKa = 5.92MAQKK74 pKa = 10.32IKK76 pKa = 10.51FGYY79 pKa = 7.95HH80 pKa = 5.87HH81 pKa = 7.42PVASLPFMITNLKK94 pKa = 10.55KK95 pKa = 10.7GDD97 pKa = 4.16LPDD100 pKa = 4.13HH101 pKa = 6.57PVPHH105 pKa = 6.86DD106 pKa = 3.4QHH108 pKa = 5.34YY109 pKa = 7.47TAARR113 pKa = 11.84KK114 pKa = 8.69AAADD118 pKa = 3.4AFRR121 pKa = 11.84PPRR124 pKa = 11.84LVRR127 pKa = 11.84PVHH130 pKa = 5.99FADD133 pKa = 3.66LRR135 pKa = 11.84YY136 pKa = 10.32YY137 pKa = 9.43KK138 pKa = 10.13WNWHH142 pKa = 6.31PNVEE146 pKa = 4.18EE147 pKa = 4.93PYY149 pKa = 10.86YY150 pKa = 10.76SDD152 pKa = 4.85PKK154 pKa = 8.47LQQYY158 pKa = 9.07VEE160 pKa = 3.99HH161 pKa = 7.34CYY163 pKa = 11.07ALGLIDD169 pKa = 5.33DD170 pKa = 4.7ARR172 pKa = 11.84LSFGNLKK179 pKa = 10.76DD180 pKa = 3.91FVFMDD185 pKa = 3.51TRR187 pKa = 11.84HH188 pKa = 5.57YY189 pKa = 11.06LHH191 pKa = 7.36LIKK194 pKa = 10.87NGSITDD200 pKa = 3.89NNQLWPIMKK209 pKa = 9.55IHH211 pKa = 6.55VKK213 pKa = 9.79PALTEE218 pKa = 3.8PTEE221 pKa = 4.01TKK223 pKa = 9.93IRR225 pKa = 11.84VIYY228 pKa = 9.95GVSKK232 pKa = 10.58RR233 pKa = 11.84HH234 pKa = 6.15ILAQAMFFWPLFRR247 pKa = 11.84YY248 pKa = 9.91YY249 pKa = 9.75IEE251 pKa = 4.36EE252 pKa = 4.16HH253 pKa = 6.1TSPLLWGNEE262 pKa = 3.87TFTGGMLKK270 pKa = 9.36IHH272 pKa = 6.27NLISVPRR279 pKa = 11.84LYY281 pKa = 10.81SQTYY285 pKa = 8.02LTVDD289 pKa = 3.2WSGFDD294 pKa = 3.54LRR296 pKa = 11.84SLFTIQRR303 pKa = 11.84EE304 pKa = 4.15IFDD307 pKa = 3.37DD308 pKa = 2.94WRR310 pKa = 11.84TYY312 pKa = 11.32FDD314 pKa = 3.47FTAYY318 pKa = 9.66IPTRR322 pKa = 11.84TYY324 pKa = 10.71PDD326 pKa = 3.51SKK328 pKa = 9.65TDD330 pKa = 4.41PIRR333 pKa = 11.84MEE335 pKa = 5.12RR336 pKa = 11.84LWNWQRR342 pKa = 11.84DD343 pKa = 3.86ACFKK347 pKa = 10.6MPFVLPDD354 pKa = 3.04RR355 pKa = 11.84TTYY358 pKa = 11.13ARR360 pKa = 11.84LFRR363 pKa = 11.84SIPSGLFVTQFLDD376 pKa = 3.27SHH378 pKa = 6.99YY379 pKa = 9.9NLIMIYY385 pKa = 9.88TILSAMGFDD394 pKa = 3.64ITNLMILVQGDD405 pKa = 3.69DD406 pKa = 3.75SLIHH410 pKa = 6.62LKK412 pKa = 10.59FFLPADD418 pKa = 3.26QHH420 pKa = 6.72DD421 pKa = 4.03AFKK424 pKa = 11.49AEE426 pKa = 4.15FEE428 pKa = 4.29RR429 pKa = 11.84LAKK432 pKa = 10.7YY433 pKa = 10.63YY434 pKa = 9.68FDD436 pKa = 5.56HH437 pKa = 7.21IARR440 pKa = 11.84PEE442 pKa = 4.02KK443 pKa = 9.8THH445 pKa = 5.29VTNSPNEE452 pKa = 4.07VEE454 pKa = 4.28VLGYY458 pKa = 9.19TNNNGYY464 pKa = 9.58PSRR467 pKa = 11.84DD468 pKa = 3.2MTKK471 pKa = 10.55LVAQLYY477 pKa = 9.88HH478 pKa = 6.62PRR480 pKa = 11.84NVDD483 pKa = 3.01KK484 pKa = 10.98TSWKK488 pKa = 9.47SLLMAKK494 pKa = 9.82VCGFAYY500 pKa = 10.29ASCYY504 pKa = 9.75QDD506 pKa = 3.06SQVIDD511 pKa = 4.18LLRR514 pKa = 11.84SIYY517 pKa = 10.99NNLASKK523 pKa = 10.39GFKK526 pKa = 9.49PKK528 pKa = 9.97SGRR531 pKa = 11.84VMRR534 pKa = 11.84DD535 pKa = 2.66IILFGEE541 pKa = 4.59SEE543 pKa = 4.55FEE545 pKa = 4.17VPTDD549 pKa = 4.18HH550 pKa = 7.52FPTLNDD556 pKa = 2.85VTKK559 pKa = 10.09YY560 pKa = 9.69FRR562 pKa = 11.84RR563 pKa = 11.84PYY565 pKa = 10.82VRR567 pKa = 11.84TQRR570 pKa = 11.84DD571 pKa = 3.12ADD573 pKa = 3.77SYY575 pKa = 10.3FPSWHH580 pKa = 6.42FNDD583 pKa = 3.7VFF585 pKa = 4.39
Molecular weight: 68.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1106 |
521 |
585 |
553.0 |
62.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.595 ± 1.685 | 0.814 ± 0.103 |
5.967 ± 0.961 | 3.074 ± 0.273 |
6.781 ± 0.587 | 5.154 ± 0.834 |
3.255 ± 0.944 | 4.882 ± 0.602 |
4.611 ± 1.088 | 8.951 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.712 ± 0.118 | 5.335 ± 0.977 |
6.51 ± 0.282 | 4.069 ± 0.787 |
4.973 ± 0.53 | 7.233 ± 1.127 |
5.877 ± 0.22 | 5.696 ± 0.451 |
1.627 ± 0.336 | 4.882 ± 1.144 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
