
Ictalurid herpesvirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Ictalurivirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
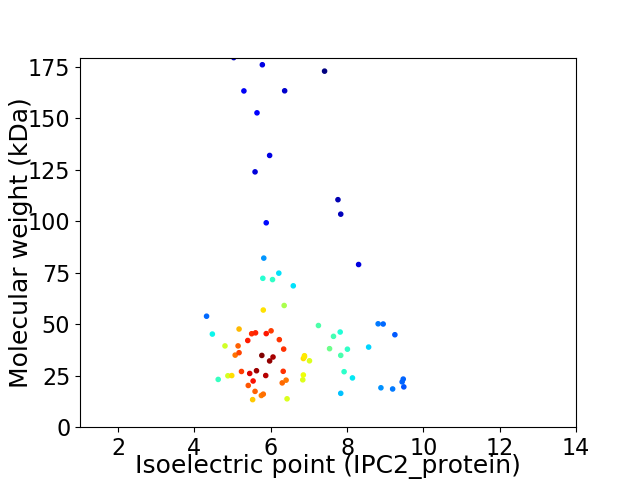
Virtual 2D-PAGE plot for 77 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

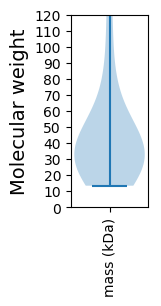
Protein with the lowest isoelectric point:
>tr|A0A2H5AJF0|A0A2H5AJF0_9VIRU ORF11L OS=Ictalurid herpesvirus 2 OX=508441 PE=4 SV=1
MM1 pKa = 7.62ASNMARR7 pKa = 11.84AGHH10 pKa = 5.16VHH12 pKa = 6.3WCLQLFSEE20 pKa = 5.28LGPRR24 pKa = 11.84DD25 pKa = 4.16DD26 pKa = 5.1DD27 pKa = 3.92QSSSSSSSSGSSVSGDD43 pKa = 3.36TPDD46 pKa = 3.06WVSTILEE53 pKa = 4.26TSDD56 pKa = 5.47DD57 pKa = 4.69DD58 pKa = 4.71LSSMPSPVSDD68 pKa = 3.33RR69 pKa = 11.84TSSAFSWASDD79 pKa = 3.9DD80 pKa = 5.0EE81 pKa = 4.91EE82 pKa = 6.96GDD84 pKa = 3.7DD85 pKa = 5.45DD86 pKa = 4.85VFEE89 pKa = 4.52AAPGGSWSEE98 pKa = 3.75RR99 pKa = 11.84TPAEE103 pKa = 4.1VEE105 pKa = 3.85VRR107 pKa = 11.84TPGSSDD113 pKa = 3.87DD114 pKa = 3.83EE115 pKa = 4.89VVCLGVVEE123 pKa = 4.3KK124 pKa = 11.05VRR126 pKa = 11.84LVRR129 pKa = 11.84EE130 pKa = 4.1GGVWRR135 pKa = 11.84VAEE138 pKa = 4.18RR139 pKa = 11.84RR140 pKa = 11.84VEE142 pKa = 4.22APTPYY147 pKa = 9.92PWANPEE153 pKa = 3.97EE154 pKa = 4.33PTAEE158 pKa = 4.03DD159 pKa = 4.42FFTEE163 pKa = 4.45FAEE166 pKa = 4.48FDD168 pKa = 3.89EE169 pKa = 5.2LDD171 pKa = 3.62EE172 pKa = 4.43LAAAIANTPGSPSQYY187 pKa = 10.11SPPSPDD193 pKa = 3.43PSHH196 pKa = 7.89HH197 pKa = 6.32EE198 pKa = 4.15DD199 pKa = 3.61EE200 pKa = 5.04YY201 pKa = 11.4EE202 pKa = 4.21NPPSPTDD209 pKa = 3.58YY210 pKa = 11.16SPTSPRR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.23SLDD221 pKa = 3.48EE222 pKa = 4.0EE223 pKa = 4.54APAYY227 pKa = 10.78SPITSPEE234 pKa = 3.25WDD236 pKa = 3.71YY237 pKa = 10.65EE238 pKa = 4.31TRR240 pKa = 11.84ASPRR244 pKa = 11.84APEE247 pKa = 3.94VAEE250 pKa = 4.14VAEE253 pKa = 4.3VAPTRR258 pKa = 11.84GFHH261 pKa = 5.91MGPLLSDD268 pKa = 3.24AWVSYY273 pKa = 10.86EE274 pKa = 4.06EE275 pKa = 4.18YY276 pKa = 11.23ANDD279 pKa = 4.5FIMDD283 pKa = 3.75YY284 pKa = 9.29RR285 pKa = 11.84TLYY288 pKa = 9.95HH289 pKa = 5.99PHH291 pKa = 7.52RR292 pKa = 11.84DD293 pKa = 3.6TPYY296 pKa = 10.68SPDD299 pKa = 3.02EE300 pKa = 4.16RR301 pKa = 11.84RR302 pKa = 11.84AGEE305 pKa = 4.39TPHH308 pKa = 6.81GPYY311 pKa = 10.47VPDD314 pKa = 3.87SPVAGPSRR322 pKa = 11.84KK323 pKa = 9.26RR324 pKa = 11.84GRR326 pKa = 11.84KK327 pKa = 9.67GEE329 pKa = 4.3TKK331 pKa = 10.36GAPPKK336 pKa = 10.47KK337 pKa = 9.6KK338 pKa = 10.43ALVEE342 pKa = 4.16GWDD345 pKa = 3.82EE346 pKa = 3.89DD347 pKa = 3.92WEE349 pKa = 5.64AEE351 pKa = 4.11LQRR354 pKa = 11.84IRR356 pKa = 11.84DD357 pKa = 4.14GYY359 pKa = 9.66MGTPITRR366 pKa = 11.84HH367 pKa = 6.44DD368 pKa = 4.44LLPATVEE375 pKa = 3.59ICPISNCFFSGLKK388 pKa = 10.25PEE390 pKa = 4.33INGPTVCKK398 pKa = 10.35CAICKK403 pKa = 9.91HH404 pKa = 6.59DD405 pKa = 4.06GQCLCLARR413 pKa = 11.84CARR416 pKa = 11.84NCPDD420 pKa = 3.81YY421 pKa = 11.38EE422 pKa = 4.5CTCVDD427 pKa = 2.96IATDD431 pKa = 3.56NGRR434 pKa = 11.84DD435 pKa = 3.75PNPIFDD441 pKa = 5.02PEE443 pKa = 3.95DD444 pKa = 3.42TRR446 pKa = 11.84NFFIDD451 pKa = 3.16EE452 pKa = 4.23CLAKK456 pKa = 10.79GLTDD460 pKa = 3.9EE461 pKa = 4.47VASDD465 pKa = 4.17LFTAKK470 pKa = 9.45TLAFQIRR477 pKa = 11.84NEE479 pKa = 4.04TRR481 pKa = 11.84LKK483 pKa = 10.88EE484 pKa = 3.93LDD486 pKa = 3.5NAA488 pKa = 4.47
MM1 pKa = 7.62ASNMARR7 pKa = 11.84AGHH10 pKa = 5.16VHH12 pKa = 6.3WCLQLFSEE20 pKa = 5.28LGPRR24 pKa = 11.84DD25 pKa = 4.16DD26 pKa = 5.1DD27 pKa = 3.92QSSSSSSSSGSSVSGDD43 pKa = 3.36TPDD46 pKa = 3.06WVSTILEE53 pKa = 4.26TSDD56 pKa = 5.47DD57 pKa = 4.69DD58 pKa = 4.71LSSMPSPVSDD68 pKa = 3.33RR69 pKa = 11.84TSSAFSWASDD79 pKa = 3.9DD80 pKa = 5.0EE81 pKa = 4.91EE82 pKa = 6.96GDD84 pKa = 3.7DD85 pKa = 5.45DD86 pKa = 4.85VFEE89 pKa = 4.52AAPGGSWSEE98 pKa = 3.75RR99 pKa = 11.84TPAEE103 pKa = 4.1VEE105 pKa = 3.85VRR107 pKa = 11.84TPGSSDD113 pKa = 3.87DD114 pKa = 3.83EE115 pKa = 4.89VVCLGVVEE123 pKa = 4.3KK124 pKa = 11.05VRR126 pKa = 11.84LVRR129 pKa = 11.84EE130 pKa = 4.1GGVWRR135 pKa = 11.84VAEE138 pKa = 4.18RR139 pKa = 11.84RR140 pKa = 11.84VEE142 pKa = 4.22APTPYY147 pKa = 9.92PWANPEE153 pKa = 3.97EE154 pKa = 4.33PTAEE158 pKa = 4.03DD159 pKa = 4.42FFTEE163 pKa = 4.45FAEE166 pKa = 4.48FDD168 pKa = 3.89EE169 pKa = 5.2LDD171 pKa = 3.62EE172 pKa = 4.43LAAAIANTPGSPSQYY187 pKa = 10.11SPPSPDD193 pKa = 3.43PSHH196 pKa = 7.89HH197 pKa = 6.32EE198 pKa = 4.15DD199 pKa = 3.61EE200 pKa = 5.04YY201 pKa = 11.4EE202 pKa = 4.21NPPSPTDD209 pKa = 3.58YY210 pKa = 11.16SPTSPRR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.23SLDD221 pKa = 3.48EE222 pKa = 4.0EE223 pKa = 4.54APAYY227 pKa = 10.78SPITSPEE234 pKa = 3.25WDD236 pKa = 3.71YY237 pKa = 10.65EE238 pKa = 4.31TRR240 pKa = 11.84ASPRR244 pKa = 11.84APEE247 pKa = 3.94VAEE250 pKa = 4.14VAEE253 pKa = 4.3VAPTRR258 pKa = 11.84GFHH261 pKa = 5.91MGPLLSDD268 pKa = 3.24AWVSYY273 pKa = 10.86EE274 pKa = 4.06EE275 pKa = 4.18YY276 pKa = 11.23ANDD279 pKa = 4.5FIMDD283 pKa = 3.75YY284 pKa = 9.29RR285 pKa = 11.84TLYY288 pKa = 9.95HH289 pKa = 5.99PHH291 pKa = 7.52RR292 pKa = 11.84DD293 pKa = 3.6TPYY296 pKa = 10.68SPDD299 pKa = 3.02EE300 pKa = 4.16RR301 pKa = 11.84RR302 pKa = 11.84AGEE305 pKa = 4.39TPHH308 pKa = 6.81GPYY311 pKa = 10.47VPDD314 pKa = 3.87SPVAGPSRR322 pKa = 11.84KK323 pKa = 9.26RR324 pKa = 11.84GRR326 pKa = 11.84KK327 pKa = 9.67GEE329 pKa = 4.3TKK331 pKa = 10.36GAPPKK336 pKa = 10.47KK337 pKa = 9.6KK338 pKa = 10.43ALVEE342 pKa = 4.16GWDD345 pKa = 3.82EE346 pKa = 3.89DD347 pKa = 3.92WEE349 pKa = 5.64AEE351 pKa = 4.11LQRR354 pKa = 11.84IRR356 pKa = 11.84DD357 pKa = 4.14GYY359 pKa = 9.66MGTPITRR366 pKa = 11.84HH367 pKa = 6.44DD368 pKa = 4.44LLPATVEE375 pKa = 3.59ICPISNCFFSGLKK388 pKa = 10.25PEE390 pKa = 4.33INGPTVCKK398 pKa = 10.35CAICKK403 pKa = 9.91HH404 pKa = 6.59DD405 pKa = 4.06GQCLCLARR413 pKa = 11.84CARR416 pKa = 11.84NCPDD420 pKa = 3.81YY421 pKa = 11.38EE422 pKa = 4.5CTCVDD427 pKa = 2.96IATDD431 pKa = 3.56NGRR434 pKa = 11.84DD435 pKa = 3.75PNPIFDD441 pKa = 5.02PEE443 pKa = 3.95DD444 pKa = 3.42TRR446 pKa = 11.84NFFIDD451 pKa = 3.16EE452 pKa = 4.23CLAKK456 pKa = 10.79GLTDD460 pKa = 3.9EE461 pKa = 4.47VASDD465 pKa = 4.17LFTAKK470 pKa = 9.45TLAFQIRR477 pKa = 11.84NEE479 pKa = 4.04TRR481 pKa = 11.84LKK483 pKa = 10.88EE484 pKa = 3.93LDD486 pKa = 3.5NAA488 pKa = 4.47
Molecular weight: 53.86 kDa
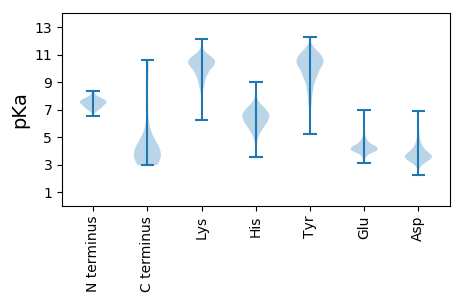
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5AJI0|A0A2H5AJI0_9VIRU ORF41 OS=Ictalurid herpesvirus 2 OX=508441 PE=4 SV=1
MM1 pKa = 7.51TFQFNCWDD9 pKa = 5.16PILDD13 pKa = 5.22DD14 pKa = 3.58ISKK17 pKa = 8.71TIQIATVDD25 pKa = 4.12RR26 pKa = 11.84QPPTYY31 pKa = 9.4WNYY34 pKa = 8.27ITAEE38 pKa = 3.88TGMRR42 pKa = 11.84GYY44 pKa = 10.84EE45 pKa = 3.92LFGYY49 pKa = 9.73PSLTQSGTIPGSPWRR64 pKa = 11.84PEE66 pKa = 3.63MSTLSWPVPATSVHH80 pKa = 6.51EE81 pKa = 4.09DD82 pKa = 3.3VIMAEE87 pKa = 4.32PKK89 pKa = 10.59VPIQPRR95 pKa = 11.84QHH97 pKa = 5.73RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84SRR104 pKa = 11.84GAGRR108 pKa = 11.84KK109 pKa = 8.68SVHH112 pKa = 6.19ALAPVWSTPSPGTAVRR128 pKa = 11.84QVPQEE133 pKa = 3.93QRR135 pKa = 11.84QQPPTPIKK143 pKa = 9.74TGSLKK148 pKa = 11.03SPGAPKK154 pKa = 10.24SPRR157 pKa = 11.84VKK159 pKa = 10.38RR160 pKa = 11.84GVTFADD166 pKa = 3.21ACTIVQFDD174 pKa = 4.82RR175 pKa = 11.84EE176 pKa = 4.24DD177 pKa = 3.53QPDD180 pKa = 3.94VVHH183 pKa = 6.39GAPRR187 pKa = 11.84GRR189 pKa = 11.84VALRR193 pKa = 11.84PPNLL197 pKa = 3.4
MM1 pKa = 7.51TFQFNCWDD9 pKa = 5.16PILDD13 pKa = 5.22DD14 pKa = 3.58ISKK17 pKa = 8.71TIQIATVDD25 pKa = 4.12RR26 pKa = 11.84QPPTYY31 pKa = 9.4WNYY34 pKa = 8.27ITAEE38 pKa = 3.88TGMRR42 pKa = 11.84GYY44 pKa = 10.84EE45 pKa = 3.92LFGYY49 pKa = 9.73PSLTQSGTIPGSPWRR64 pKa = 11.84PEE66 pKa = 3.63MSTLSWPVPATSVHH80 pKa = 6.51EE81 pKa = 4.09DD82 pKa = 3.3VIMAEE87 pKa = 4.32PKK89 pKa = 10.59VPIQPRR95 pKa = 11.84QHH97 pKa = 5.73RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84SRR104 pKa = 11.84GAGRR108 pKa = 11.84KK109 pKa = 8.68SVHH112 pKa = 6.19ALAPVWSTPSPGTAVRR128 pKa = 11.84QVPQEE133 pKa = 3.93QRR135 pKa = 11.84QQPPTPIKK143 pKa = 9.74TGSLKK148 pKa = 11.03SPGAPKK154 pKa = 10.24SPRR157 pKa = 11.84VKK159 pKa = 10.38RR160 pKa = 11.84GVTFADD166 pKa = 3.21ACTIVQFDD174 pKa = 4.82RR175 pKa = 11.84EE176 pKa = 4.24DD177 pKa = 3.53QPDD180 pKa = 3.94VVHH183 pKa = 6.39GAPRR187 pKa = 11.84GRR189 pKa = 11.84VALRR193 pKa = 11.84PPNLL197 pKa = 3.4
Molecular weight: 21.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35900 |
120 |
1629 |
466.2 |
51.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.12 ± 0.198 | 1.71 ± 0.131 |
5.457 ± 0.15 | 6.209 ± 0.28 |
3.836 ± 0.178 | 7.061 ± 0.154 |
2.32 ± 0.098 | 5.855 ± 0.199 |
4.591 ± 0.266 | 9.365 ± 0.197 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.922 ± 0.107 | 3.454 ± 0.131 |
6.17 ± 0.34 | 2.986 ± 0.137 |
6.345 ± 0.215 | 6.003 ± 0.209 |
7.437 ± 0.181 | 6.981 ± 0.127 |
1.276 ± 0.088 | 2.903 ± 0.143 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
