
Verticillium alfalfae (strain VaMs.102 / ATCC MYA-4576 / FGSC 10136) (Verticillium wilt of alfalfa) (Verticillium albo-atrum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Glomerellales; Plectosphaerellaceae; Verticillium; Verticillium alfalfae
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
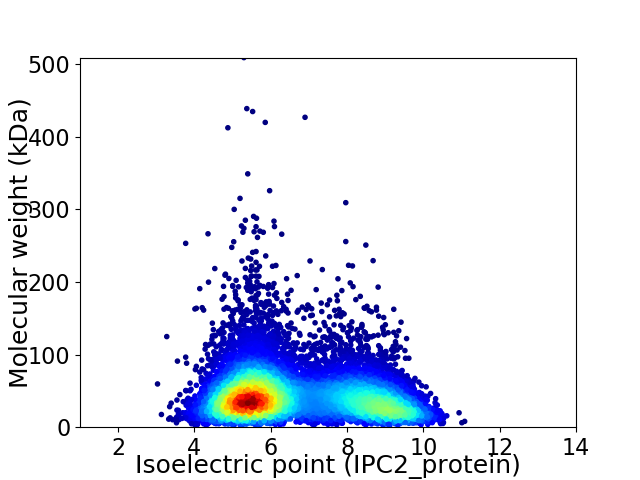
Virtual 2D-PAGE plot for 10233 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C9SNW9|C9SNW9_VERA1 Parasitic phase-specific protein PSP-1 OS=Verticillium alfalfae (strain VaMs.102 / ATCC MYA-4576 / FGSC 10136) OX=526221 GN=VDBG_06594 PE=4 SV=1
MM1 pKa = 6.68VASYY5 pKa = 9.58TKK7 pKa = 10.68LAIALTAASNVVAFTSIRR25 pKa = 11.84IAPTVEE31 pKa = 3.49AGKK34 pKa = 9.74EE35 pKa = 4.07VEE37 pKa = 4.1VSIVNDD43 pKa = 3.83LADD46 pKa = 4.17SDD48 pKa = 4.74SFDD51 pKa = 3.71AGFEE55 pKa = 3.98KK56 pKa = 10.6FRR58 pKa = 11.84VYY60 pKa = 10.82LATTPPGWGTGPACHH75 pKa = 6.82LVNATAIDD83 pKa = 3.81EE84 pKa = 4.5TSVKK88 pKa = 8.84VTIPPSVVPDD98 pKa = 4.01GSDD101 pKa = 3.2VMISVMEE108 pKa = 4.24FNEE111 pKa = 4.5DD112 pKa = 3.39PSLDD116 pKa = 3.86GPSGFQYY123 pKa = 11.17SNEE126 pKa = 3.78FTLNGGKK133 pKa = 10.31GEE135 pKa = 4.0WSKK138 pKa = 11.67PEE140 pKa = 4.0LEE142 pKa = 4.44GFNVGDD148 pKa = 4.32SDD150 pKa = 4.94TIPCEE155 pKa = 3.98AYY157 pKa = 10.23ACARR161 pKa = 11.84DD162 pKa = 4.14CMVQFYY168 pKa = 10.16PDD170 pKa = 3.44NKK172 pKa = 9.61EE173 pKa = 4.13DD174 pKa = 3.29EE175 pKa = 4.24SAYY178 pKa = 10.61RR179 pKa = 11.84KK180 pKa = 8.42TYY182 pKa = 10.26EE183 pKa = 4.5CTAEE187 pKa = 4.34CPGVDD192 pKa = 3.71YY193 pKa = 9.49PAWDD197 pKa = 3.94SLPGADD203 pKa = 4.38EE204 pKa = 4.48VPSEE208 pKa = 5.25DD209 pKa = 5.48DD210 pKa = 4.77GDD212 pKa = 5.17DD213 pKa = 3.92EE214 pKa = 7.46GDD216 pKa = 4.72DD217 pKa = 4.19DD218 pKa = 6.67DD219 pKa = 4.28EE220 pKa = 4.51TASISAAPSSIKK232 pKa = 9.98TSAATSGTATPSSGAATTILTTTGADD258 pKa = 3.21EE259 pKa = 4.75ASAPTEE265 pKa = 4.19TPSAAGRR272 pKa = 11.84HH273 pKa = 4.83TASLMLLAGIIGMCCLFF290 pKa = 4.34
MM1 pKa = 6.68VASYY5 pKa = 9.58TKK7 pKa = 10.68LAIALTAASNVVAFTSIRR25 pKa = 11.84IAPTVEE31 pKa = 3.49AGKK34 pKa = 9.74EE35 pKa = 4.07VEE37 pKa = 4.1VSIVNDD43 pKa = 3.83LADD46 pKa = 4.17SDD48 pKa = 4.74SFDD51 pKa = 3.71AGFEE55 pKa = 3.98KK56 pKa = 10.6FRR58 pKa = 11.84VYY60 pKa = 10.82LATTPPGWGTGPACHH75 pKa = 6.82LVNATAIDD83 pKa = 3.81EE84 pKa = 4.5TSVKK88 pKa = 8.84VTIPPSVVPDD98 pKa = 4.01GSDD101 pKa = 3.2VMISVMEE108 pKa = 4.24FNEE111 pKa = 4.5DD112 pKa = 3.39PSLDD116 pKa = 3.86GPSGFQYY123 pKa = 11.17SNEE126 pKa = 3.78FTLNGGKK133 pKa = 10.31GEE135 pKa = 4.0WSKK138 pKa = 11.67PEE140 pKa = 4.0LEE142 pKa = 4.44GFNVGDD148 pKa = 4.32SDD150 pKa = 4.94TIPCEE155 pKa = 3.98AYY157 pKa = 10.23ACARR161 pKa = 11.84DD162 pKa = 4.14CMVQFYY168 pKa = 10.16PDD170 pKa = 3.44NKK172 pKa = 9.61EE173 pKa = 4.13DD174 pKa = 3.29EE175 pKa = 4.24SAYY178 pKa = 10.61RR179 pKa = 11.84KK180 pKa = 8.42TYY182 pKa = 10.26EE183 pKa = 4.5CTAEE187 pKa = 4.34CPGVDD192 pKa = 3.71YY193 pKa = 9.49PAWDD197 pKa = 3.94SLPGADD203 pKa = 4.38EE204 pKa = 4.48VPSEE208 pKa = 5.25DD209 pKa = 5.48DD210 pKa = 4.77GDD212 pKa = 5.17DD213 pKa = 3.92EE214 pKa = 7.46GDD216 pKa = 4.72DD217 pKa = 4.19DD218 pKa = 6.67DD219 pKa = 4.28EE220 pKa = 4.51TASISAAPSSIKK232 pKa = 9.98TSAATSGTATPSSGAATTILTTTGADD258 pKa = 3.21EE259 pKa = 4.75ASAPTEE265 pKa = 4.19TPSAAGRR272 pKa = 11.84HH273 pKa = 4.83TASLMLLAGIIGMCCLFF290 pKa = 4.34
Molecular weight: 30.18 kDa
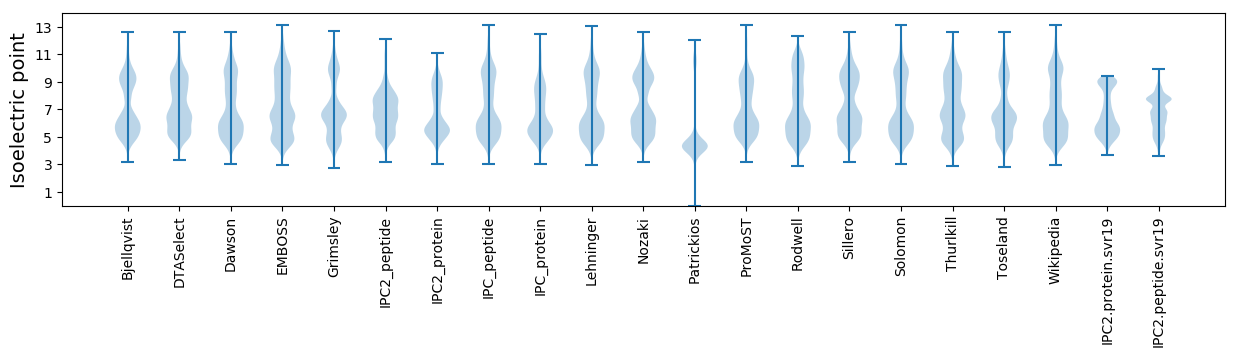
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9SYL7|C9SYL7_VERA1 Decarboxylase DEC1 OS=Verticillium alfalfae (strain VaMs.102 / ATCC MYA-4576 / FGSC 10136) OX=526221 GN=VDBG_09992 PE=4 SV=1
MM1 pKa = 7.56SFRR4 pKa = 11.84GGRR7 pKa = 11.84GAPRR11 pKa = 11.84GFGRR15 pKa = 11.84SWVEE19 pKa = 3.56AVEE22 pKa = 4.37VSSNATWVPRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.78WASSCTPARR43 pKa = 11.84VRR45 pKa = 11.84WSARR49 pKa = 11.84ASTPRR54 pKa = 11.84YY55 pKa = 8.38PSSTPRR61 pKa = 11.84CSSRR65 pKa = 11.84TRR67 pKa = 11.84PPLARR72 pKa = 11.84STKK75 pKa = 10.27SSAPSTKK82 pKa = 9.0STSPSSPPRR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84TWRR96 pKa = 11.84LRR98 pKa = 11.84RR99 pKa = 11.84SWRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84FRR107 pKa = 11.84RR108 pKa = 11.84TRR110 pKa = 11.84RR111 pKa = 11.84CAEE114 pKa = 3.25GWTRR118 pKa = 11.84RR119 pKa = 11.84LQRR122 pKa = 11.84RR123 pKa = 11.84SWRR126 pKa = 11.84QRR128 pKa = 11.84LWWSRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84LWRR140 pKa = 11.84SRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84AEE146 pKa = 3.89GRR148 pKa = 11.84RR149 pKa = 11.84QGTRR153 pKa = 11.84RR154 pKa = 11.84LQQGRR159 pKa = 11.84RR160 pKa = 11.84ILRR163 pKa = 11.84LL164 pKa = 3.27
MM1 pKa = 7.56SFRR4 pKa = 11.84GGRR7 pKa = 11.84GAPRR11 pKa = 11.84GFGRR15 pKa = 11.84SWVEE19 pKa = 3.56AVEE22 pKa = 4.37VSSNATWVPRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.78WASSCTPARR43 pKa = 11.84VRR45 pKa = 11.84WSARR49 pKa = 11.84ASTPRR54 pKa = 11.84YY55 pKa = 8.38PSSTPRR61 pKa = 11.84CSSRR65 pKa = 11.84TRR67 pKa = 11.84PPLARR72 pKa = 11.84STKK75 pKa = 10.27SSAPSTKK82 pKa = 9.0STSPSSPPRR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84TWRR96 pKa = 11.84LRR98 pKa = 11.84RR99 pKa = 11.84SWRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84FRR107 pKa = 11.84RR108 pKa = 11.84TRR110 pKa = 11.84RR111 pKa = 11.84CAEE114 pKa = 3.25GWTRR118 pKa = 11.84RR119 pKa = 11.84LQRR122 pKa = 11.84RR123 pKa = 11.84SWRR126 pKa = 11.84QRR128 pKa = 11.84LWWSRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84LWRR140 pKa = 11.84SRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84AEE146 pKa = 3.89GRR148 pKa = 11.84RR149 pKa = 11.84QGTRR153 pKa = 11.84RR154 pKa = 11.84LQQGRR159 pKa = 11.84RR160 pKa = 11.84ILRR163 pKa = 11.84LL164 pKa = 3.27
Molecular weight: 19.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4470746 |
29 |
4615 |
436.9 |
48.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.631 ± 0.022 | 1.263 ± 0.01 |
5.831 ± 0.016 | 5.908 ± 0.02 |
3.568 ± 0.015 | 7.2 ± 0.025 |
2.452 ± 0.011 | 4.45 ± 0.018 |
4.487 ± 0.021 | 8.688 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.186 ± 0.01 | 3.388 ± 0.014 |
6.286 ± 0.025 | 3.989 ± 0.019 |
6.513 ± 0.02 | 7.951 ± 0.028 |
5.968 ± 0.015 | 6.243 ± 0.017 |
1.474 ± 0.009 | 2.525 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
