
Measles virus (strain Ichinose-B95a) (MeV) (Subacute sclerose panencephalitis virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus; Measles morbillivirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
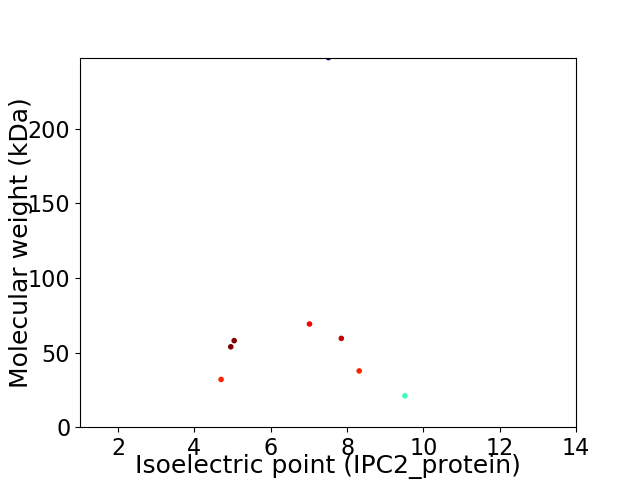
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
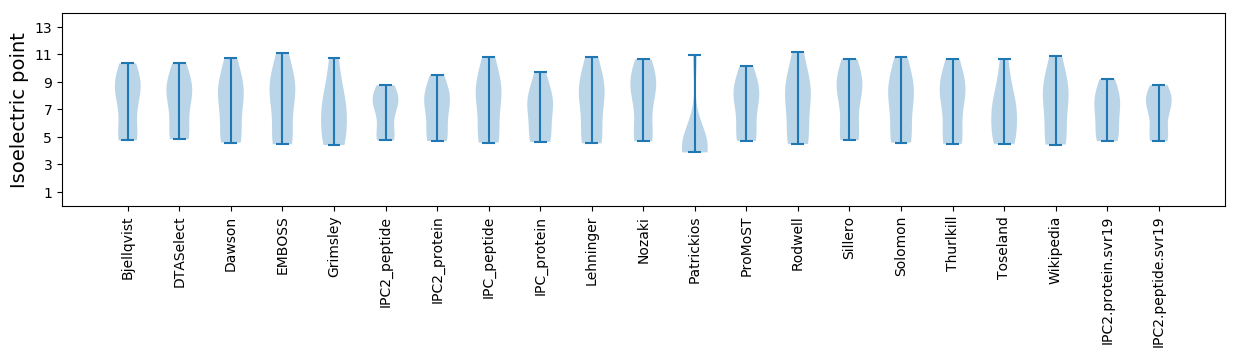
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q786F2|HEMA_MEASC Hemagglutinin glycoprotein OS=Measles virus (strain Ichinose-B95a) OX=645098 GN=H PE=1 SV=1
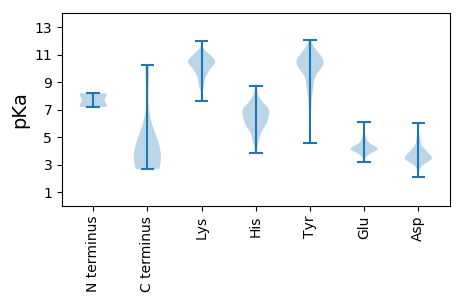
MM1 pKa = 8.15AEE3 pKa = 3.63EE4 pKa = 3.91QARR7 pKa = 11.84HH8 pKa = 4.84VKK10 pKa = 10.61NGLEE14 pKa = 4.48CIRR17 pKa = 11.84ALKK20 pKa = 10.54AEE22 pKa = 5.1PIGSLAVEE30 pKa = 4.26EE31 pKa = 5.78AMAAWSEE38 pKa = 4.06ISDD41 pKa = 3.99NPGQDD46 pKa = 2.54RR47 pKa = 11.84ATCKK51 pKa = 10.55EE52 pKa = 4.09EE53 pKa = 3.89EE54 pKa = 4.39AGSSGLSKK62 pKa = 10.51PCLSAIGSTEE72 pKa = 3.52GGAPRR77 pKa = 11.84IRR79 pKa = 11.84GQGSGEE85 pKa = 4.09SDD87 pKa = 3.97DD88 pKa = 4.82DD89 pKa = 4.92AEE91 pKa = 4.47TLGIPSRR98 pKa = 11.84NLQASSTGLQCYY110 pKa = 8.88HH111 pKa = 7.22VYY113 pKa = 10.56DD114 pKa = 4.08HH115 pKa = 7.03SGEE118 pKa = 4.13AVKK121 pKa = 10.71GIQDD125 pKa = 3.6ADD127 pKa = 3.98SIMVQSGLDD136 pKa = 3.52GDD138 pKa = 4.4STLSGGDD145 pKa = 3.82DD146 pKa = 3.42EE147 pKa = 6.08SEE149 pKa = 4.18NSDD152 pKa = 3.3VDD154 pKa = 3.07IGEE157 pKa = 4.99PDD159 pKa = 3.41TEE161 pKa = 4.81GYY163 pKa = 10.86AITDD167 pKa = 3.66RR168 pKa = 11.84GSAPISMGFRR178 pKa = 11.84ASDD181 pKa = 3.44VEE183 pKa = 4.22TAEE186 pKa = 4.19GGEE189 pKa = 3.8IHH191 pKa = 7.05EE192 pKa = 4.42LLKK195 pKa = 10.95LQSRR199 pKa = 11.84GNNFPKK205 pKa = 10.39LGKK208 pKa = 7.92TLNVPPPPNPSRR220 pKa = 11.84ASTSEE225 pKa = 3.87TPIKK229 pKa = 9.98KK230 pKa = 8.95GHH232 pKa = 5.76RR233 pKa = 11.84RR234 pKa = 11.84EE235 pKa = 5.2IGLIWNGDD243 pKa = 3.23RR244 pKa = 11.84VFIDD248 pKa = 3.78RR249 pKa = 11.84WCNPMCSKK257 pKa = 9.47VTLGTIRR264 pKa = 11.84ARR266 pKa = 11.84CTCGEE271 pKa = 4.22CPRR274 pKa = 11.84VCEE277 pKa = 4.19QCRR280 pKa = 11.84TDD282 pKa = 3.22TGVDD286 pKa = 3.22TRR288 pKa = 11.84IWYY291 pKa = 9.3HH292 pKa = 5.99NLPEE296 pKa = 5.03IPEE299 pKa = 4.01
MM1 pKa = 8.15AEE3 pKa = 3.63EE4 pKa = 3.91QARR7 pKa = 11.84HH8 pKa = 4.84VKK10 pKa = 10.61NGLEE14 pKa = 4.48CIRR17 pKa = 11.84ALKK20 pKa = 10.54AEE22 pKa = 5.1PIGSLAVEE30 pKa = 4.26EE31 pKa = 5.78AMAAWSEE38 pKa = 4.06ISDD41 pKa = 3.99NPGQDD46 pKa = 2.54RR47 pKa = 11.84ATCKK51 pKa = 10.55EE52 pKa = 4.09EE53 pKa = 3.89EE54 pKa = 4.39AGSSGLSKK62 pKa = 10.51PCLSAIGSTEE72 pKa = 3.52GGAPRR77 pKa = 11.84IRR79 pKa = 11.84GQGSGEE85 pKa = 4.09SDD87 pKa = 3.97DD88 pKa = 4.82DD89 pKa = 4.92AEE91 pKa = 4.47TLGIPSRR98 pKa = 11.84NLQASSTGLQCYY110 pKa = 8.88HH111 pKa = 7.22VYY113 pKa = 10.56DD114 pKa = 4.08HH115 pKa = 7.03SGEE118 pKa = 4.13AVKK121 pKa = 10.71GIQDD125 pKa = 3.6ADD127 pKa = 3.98SIMVQSGLDD136 pKa = 3.52GDD138 pKa = 4.4STLSGGDD145 pKa = 3.82DD146 pKa = 3.42EE147 pKa = 6.08SEE149 pKa = 4.18NSDD152 pKa = 3.3VDD154 pKa = 3.07IGEE157 pKa = 4.99PDD159 pKa = 3.41TEE161 pKa = 4.81GYY163 pKa = 10.86AITDD167 pKa = 3.66RR168 pKa = 11.84GSAPISMGFRR178 pKa = 11.84ASDD181 pKa = 3.44VEE183 pKa = 4.22TAEE186 pKa = 4.19GGEE189 pKa = 3.8IHH191 pKa = 7.05EE192 pKa = 4.42LLKK195 pKa = 10.95LQSRR199 pKa = 11.84GNNFPKK205 pKa = 10.39LGKK208 pKa = 7.92TLNVPPPPNPSRR220 pKa = 11.84ASTSEE225 pKa = 3.87TPIKK229 pKa = 9.98KK230 pKa = 8.95GHH232 pKa = 5.76RR233 pKa = 11.84RR234 pKa = 11.84EE235 pKa = 5.2IGLIWNGDD243 pKa = 3.23RR244 pKa = 11.84VFIDD248 pKa = 3.78RR249 pKa = 11.84WCNPMCSKK257 pKa = 9.47VTLGTIRR264 pKa = 11.84ARR266 pKa = 11.84CTCGEE271 pKa = 4.22CPRR274 pKa = 11.84VCEE277 pKa = 4.19QCRR280 pKa = 11.84TDD282 pKa = 3.22TGVDD286 pKa = 3.22TRR288 pKa = 11.84IWYY291 pKa = 9.3HH292 pKa = 5.99NLPEE296 pKa = 5.03IPEE299 pKa = 4.01
Molecular weight: 32.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9YZN9|C_MEASC Protein C OS=Measles virus (strain Ichinose-B95a) OX=645098 GN=P/V/C PE=3 SV=1
MM1 pKa = 7.9SKK3 pKa = 9.54TDD5 pKa = 3.3WNASGLSRR13 pKa = 11.84PSPSAHH19 pKa = 5.55WPSRR23 pKa = 11.84KK24 pKa = 8.87PWQHH28 pKa = 3.85GQKK31 pKa = 10.26YY32 pKa = 7.23QTTQDD37 pKa = 3.16RR38 pKa = 11.84TEE40 pKa = 3.99PPARR44 pKa = 11.84KK45 pKa = 8.91RR46 pKa = 11.84RR47 pKa = 11.84QAVRR51 pKa = 11.84VSANHH56 pKa = 7.14ASQQLDD62 pKa = 3.29QLKK65 pKa = 10.12AVHH68 pKa = 6.38LASAVRR74 pKa = 11.84DD75 pKa = 3.67LEE77 pKa = 4.53KK78 pKa = 11.26AMTTLKK84 pKa = 10.47LWEE87 pKa = 4.53SPQEE91 pKa = 4.11ISRR94 pKa = 11.84HH95 pKa = 4.01QALGYY100 pKa = 9.1SVIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.28SKK117 pKa = 10.12MLTLSWFNQALMVIAPSQEE136 pKa = 3.32EE137 pKa = 4.5TMNLKK142 pKa = 8.49TAMWILANLIPRR154 pKa = 11.84DD155 pKa = 4.0MLSLTGDD162 pKa = 4.77LLPSLWGSGLLMLKK176 pKa = 9.76LQKK179 pKa = 10.13EE180 pKa = 4.71GRR182 pKa = 11.84STSSS186 pKa = 2.66
MM1 pKa = 7.9SKK3 pKa = 9.54TDD5 pKa = 3.3WNASGLSRR13 pKa = 11.84PSPSAHH19 pKa = 5.55WPSRR23 pKa = 11.84KK24 pKa = 8.87PWQHH28 pKa = 3.85GQKK31 pKa = 10.26YY32 pKa = 7.23QTTQDD37 pKa = 3.16RR38 pKa = 11.84TEE40 pKa = 3.99PPARR44 pKa = 11.84KK45 pKa = 8.91RR46 pKa = 11.84RR47 pKa = 11.84QAVRR51 pKa = 11.84VSANHH56 pKa = 7.14ASQQLDD62 pKa = 3.29QLKK65 pKa = 10.12AVHH68 pKa = 6.38LASAVRR74 pKa = 11.84DD75 pKa = 3.67LEE77 pKa = 4.53KK78 pKa = 11.26AMTTLKK84 pKa = 10.47LWEE87 pKa = 4.53SPQEE91 pKa = 4.11ISRR94 pKa = 11.84HH95 pKa = 4.01QALGYY100 pKa = 9.1SVIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.28SKK117 pKa = 10.12MLTLSWFNQALMVIAPSQEE136 pKa = 3.32EE137 pKa = 4.5TMNLKK142 pKa = 8.49TAMWILANLIPRR154 pKa = 11.84DD155 pKa = 4.0MLSLTGDD162 pKa = 4.77LLPSLWGSGLLMLKK176 pKa = 9.76LQKK179 pKa = 10.13EE180 pKa = 4.71GRR182 pKa = 11.84STSSS186 pKa = 2.66
Molecular weight: 21.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5202 |
186 |
2183 |
650.3 |
72.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.209 ± 0.579 | 1.788 ± 0.307 |
5.536 ± 0.429 | 5.709 ± 0.482 |
3.114 ± 0.529 | 7.017 ± 0.766 |
2.345 ± 0.46 | 7.132 ± 0.422 |
4.998 ± 0.347 | 10.554 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.499 ± 0.178 | 4.191 ± 0.289 |
4.748 ± 0.349 | 3.479 ± 0.378 |
6.075 ± 0.335 | 8.612 ± 0.505 |
5.498 ± 0.12 | 6.19 ± 0.521 |
1.096 ± 0.186 | 3.21 ± 0.493 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
