
Plasmodium falciparum MaliPS096_E11
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Laverania); Plasmodium falciparum
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
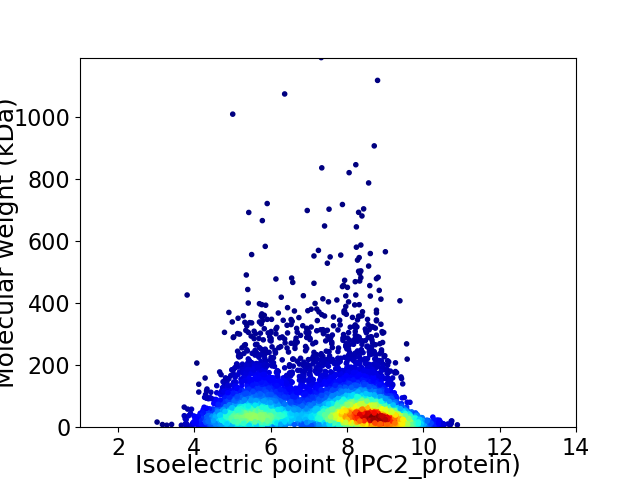
Virtual 2D-PAGE plot for 6301 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024WTS0|A0A024WTS0_PLAFA Uncharacterized protein OS=Plasmodium falciparum MaliPS096_E11 OX=1036727 GN=PFMALIP_01815 PE=4 SV=1
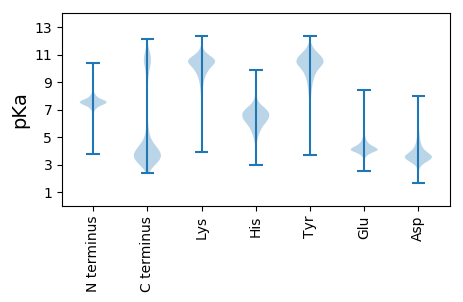
MM1 pKa = 7.67IIFKK5 pKa = 10.23RR6 pKa = 11.84YY7 pKa = 10.07HH8 pKa = 5.89NIKK11 pKa = 10.4NNNCISFSDD20 pKa = 3.93YY21 pKa = 10.72EE22 pKa = 4.08RR23 pKa = 11.84SIKK26 pKa = 10.51NFSISSHH33 pKa = 6.56AEE35 pKa = 3.52NNYY38 pKa = 10.85DD39 pKa = 4.96NIINEE44 pKa = 4.05YY45 pKa = 10.83KK46 pKa = 10.35KK47 pKa = 10.82IKK49 pKa = 10.25DD50 pKa = 3.18INNNINILSSVHH62 pKa = 5.32RR63 pKa = 11.84KK64 pKa = 9.39GRR66 pKa = 11.84ILYY69 pKa = 10.44DD70 pKa = 3.23SFLEE74 pKa = 4.26INKK77 pKa = 9.96LEE79 pKa = 3.98NDD81 pKa = 3.44KK82 pKa = 11.25KK83 pKa = 10.63EE84 pKa = 3.95KK85 pKa = 10.29HH86 pKa = 5.8EE87 pKa = 5.03KK88 pKa = 9.08EE89 pKa = 5.05DD90 pKa = 3.63EE91 pKa = 4.3YY92 pKa = 11.41EE93 pKa = 4.79DD94 pKa = 3.75NDD96 pKa = 3.54EE97 pKa = 4.82SFLEE101 pKa = 4.27TEE103 pKa = 4.36EE104 pKa = 5.43YY105 pKa = 10.63EE106 pKa = 5.04DD107 pKa = 3.65NDD109 pKa = 3.4EE110 pKa = 4.88SFLEE114 pKa = 4.27TEE116 pKa = 4.31EE117 pKa = 4.96YY118 pKa = 10.67EE119 pKa = 4.38DD120 pKa = 5.87NEE122 pKa = 4.17DD123 pKa = 3.12EE124 pKa = 5.44KK125 pKa = 11.7YY126 pKa = 11.23NKK128 pKa = 10.66DD129 pKa = 3.3EE130 pKa = 4.73DD131 pKa = 4.72DD132 pKa = 3.82YY133 pKa = 12.03AEE135 pKa = 4.56SFIEE139 pKa = 3.89TDD141 pKa = 3.16EE142 pKa = 4.8YY143 pKa = 11.26EE144 pKa = 4.63DD145 pKa = 5.45NEE147 pKa = 4.08DD148 pKa = 3.71DD149 pKa = 5.58KK150 pKa = 11.96YY151 pKa = 11.9NKK153 pKa = 10.66DD154 pKa = 3.3EE155 pKa = 4.87DD156 pKa = 4.61DD157 pKa = 3.87YY158 pKa = 11.83SEE160 pKa = 5.04SFIEE164 pKa = 3.93TDD166 pKa = 3.1EE167 pKa = 4.55YY168 pKa = 11.52DD169 pKa = 3.9DD170 pKa = 5.5NEE172 pKa = 4.04EE173 pKa = 3.73EE174 pKa = 4.76QYY176 pKa = 11.76NKK178 pKa = 11.05DD179 pKa = 3.38EE180 pKa = 5.19DD181 pKa = 4.78DD182 pKa = 4.05YY183 pKa = 11.83ADD185 pKa = 4.26SFIEE189 pKa = 3.97TDD191 pKa = 3.06HH192 pKa = 7.24YY193 pKa = 11.51EE194 pKa = 4.24NNDD197 pKa = 3.55DD198 pKa = 3.99KK199 pKa = 11.85NEE201 pKa = 3.94EE202 pKa = 3.99EE203 pKa = 4.99EE204 pKa = 4.86EE205 pKa = 4.67YY206 pKa = 11.24NDD208 pKa = 3.48QDD210 pKa = 3.52NDD212 pKa = 3.23YY213 pKa = 10.63GYY215 pKa = 11.45NFLEE219 pKa = 4.06TDD221 pKa = 3.53EE222 pKa = 5.23YY223 pKa = 11.47DD224 pKa = 3.88DD225 pKa = 4.98SEE227 pKa = 4.85EE228 pKa = 3.89YY229 pKa = 10.55DD230 pKa = 3.93YY231 pKa = 11.81DD232 pKa = 3.85DD233 pKa = 4.63KK234 pKa = 11.76EE235 pKa = 4.35YY236 pKa = 11.54GEE238 pKa = 4.57SFLEE242 pKa = 4.63KK243 pKa = 10.98EE244 pKa = 4.55EE245 pKa = 4.74DD246 pKa = 3.7DD247 pKa = 4.18EE248 pKa = 5.6AAMTAEE254 pKa = 3.98EE255 pKa = 4.57LVEE258 pKa = 4.21LEE260 pKa = 3.97NTEE263 pKa = 4.55DD264 pKa = 3.71VNTPTMVEE272 pKa = 4.08TEE274 pKa = 4.75EE275 pKa = 4.58IDD277 pKa = 3.94SDD279 pKa = 4.02EE280 pKa = 4.7NGNKK284 pKa = 9.75SSSSISYY291 pKa = 9.29ISSIVFLMVTLLYY304 pKa = 10.86FMNN307 pKa = 4.61
MM1 pKa = 7.67IIFKK5 pKa = 10.23RR6 pKa = 11.84YY7 pKa = 10.07HH8 pKa = 5.89NIKK11 pKa = 10.4NNNCISFSDD20 pKa = 3.93YY21 pKa = 10.72EE22 pKa = 4.08RR23 pKa = 11.84SIKK26 pKa = 10.51NFSISSHH33 pKa = 6.56AEE35 pKa = 3.52NNYY38 pKa = 10.85DD39 pKa = 4.96NIINEE44 pKa = 4.05YY45 pKa = 10.83KK46 pKa = 10.35KK47 pKa = 10.82IKK49 pKa = 10.25DD50 pKa = 3.18INNNINILSSVHH62 pKa = 5.32RR63 pKa = 11.84KK64 pKa = 9.39GRR66 pKa = 11.84ILYY69 pKa = 10.44DD70 pKa = 3.23SFLEE74 pKa = 4.26INKK77 pKa = 9.96LEE79 pKa = 3.98NDD81 pKa = 3.44KK82 pKa = 11.25KK83 pKa = 10.63EE84 pKa = 3.95KK85 pKa = 10.29HH86 pKa = 5.8EE87 pKa = 5.03KK88 pKa = 9.08EE89 pKa = 5.05DD90 pKa = 3.63EE91 pKa = 4.3YY92 pKa = 11.41EE93 pKa = 4.79DD94 pKa = 3.75NDD96 pKa = 3.54EE97 pKa = 4.82SFLEE101 pKa = 4.27TEE103 pKa = 4.36EE104 pKa = 5.43YY105 pKa = 10.63EE106 pKa = 5.04DD107 pKa = 3.65NDD109 pKa = 3.4EE110 pKa = 4.88SFLEE114 pKa = 4.27TEE116 pKa = 4.31EE117 pKa = 4.96YY118 pKa = 10.67EE119 pKa = 4.38DD120 pKa = 5.87NEE122 pKa = 4.17DD123 pKa = 3.12EE124 pKa = 5.44KK125 pKa = 11.7YY126 pKa = 11.23NKK128 pKa = 10.66DD129 pKa = 3.3EE130 pKa = 4.73DD131 pKa = 4.72DD132 pKa = 3.82YY133 pKa = 12.03AEE135 pKa = 4.56SFIEE139 pKa = 3.89TDD141 pKa = 3.16EE142 pKa = 4.8YY143 pKa = 11.26EE144 pKa = 4.63DD145 pKa = 5.45NEE147 pKa = 4.08DD148 pKa = 3.71DD149 pKa = 5.58KK150 pKa = 11.96YY151 pKa = 11.9NKK153 pKa = 10.66DD154 pKa = 3.3EE155 pKa = 4.87DD156 pKa = 4.61DD157 pKa = 3.87YY158 pKa = 11.83SEE160 pKa = 5.04SFIEE164 pKa = 3.93TDD166 pKa = 3.1EE167 pKa = 4.55YY168 pKa = 11.52DD169 pKa = 3.9DD170 pKa = 5.5NEE172 pKa = 4.04EE173 pKa = 3.73EE174 pKa = 4.76QYY176 pKa = 11.76NKK178 pKa = 11.05DD179 pKa = 3.38EE180 pKa = 5.19DD181 pKa = 4.78DD182 pKa = 4.05YY183 pKa = 11.83ADD185 pKa = 4.26SFIEE189 pKa = 3.97TDD191 pKa = 3.06HH192 pKa = 7.24YY193 pKa = 11.51EE194 pKa = 4.24NNDD197 pKa = 3.55DD198 pKa = 3.99KK199 pKa = 11.85NEE201 pKa = 3.94EE202 pKa = 3.99EE203 pKa = 4.99EE204 pKa = 4.86EE205 pKa = 4.67YY206 pKa = 11.24NDD208 pKa = 3.48QDD210 pKa = 3.52NDD212 pKa = 3.23YY213 pKa = 10.63GYY215 pKa = 11.45NFLEE219 pKa = 4.06TDD221 pKa = 3.53EE222 pKa = 5.23YY223 pKa = 11.47DD224 pKa = 3.88DD225 pKa = 4.98SEE227 pKa = 4.85EE228 pKa = 3.89YY229 pKa = 10.55DD230 pKa = 3.93YY231 pKa = 11.81DD232 pKa = 3.85DD233 pKa = 4.63KK234 pKa = 11.76EE235 pKa = 4.35YY236 pKa = 11.54GEE238 pKa = 4.57SFLEE242 pKa = 4.63KK243 pKa = 10.98EE244 pKa = 4.55EE245 pKa = 4.74DD246 pKa = 3.7DD247 pKa = 4.18EE248 pKa = 5.6AAMTAEE254 pKa = 3.98EE255 pKa = 4.57LVEE258 pKa = 4.21LEE260 pKa = 3.97NTEE263 pKa = 4.55DD264 pKa = 3.71VNTPTMVEE272 pKa = 4.08TEE274 pKa = 4.75EE275 pKa = 4.58IDD277 pKa = 3.94SDD279 pKa = 4.02EE280 pKa = 4.7NGNKK284 pKa = 9.75SSSSISYY291 pKa = 9.29ISSIVFLMVTLLYY304 pKa = 10.86FMNN307 pKa = 4.61
Molecular weight: 36.95 kDa
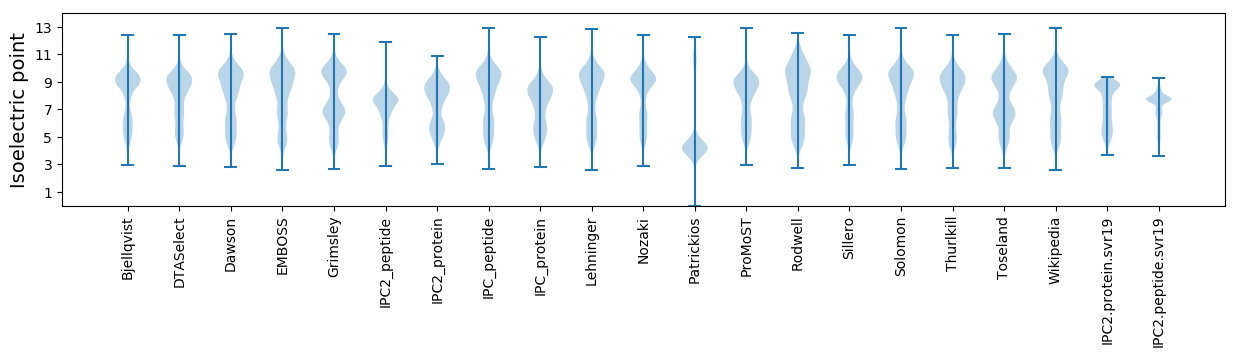
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024WV09|A0A024WV09_PLAFA Uncharacterized protein OS=Plasmodium falciparum MaliPS096_E11 OX=1036727 GN=PFMALIP_01150 PE=3 SV=1
MM1 pKa = 7.61VIRR4 pKa = 11.84EE5 pKa = 4.44SVSRR9 pKa = 11.84IYY11 pKa = 10.84VGNLPSHH18 pKa = 6.2VSSRR22 pKa = 11.84DD23 pKa = 3.23VEE25 pKa = 4.32NEE27 pKa = 3.08FRR29 pKa = 11.84KK30 pKa = 10.51YY31 pKa = 11.3GNILKK36 pKa = 10.22CDD38 pKa = 3.51VKK40 pKa = 10.59KK41 pKa = 9.2TVSGAAFAFIEE52 pKa = 4.59FEE54 pKa = 4.33DD55 pKa = 4.29ARR57 pKa = 11.84DD58 pKa = 3.3AADD61 pKa = 4.76AIKK64 pKa = 10.62EE65 pKa = 3.83KK66 pKa = 10.72DD67 pKa = 3.39GCDD70 pKa = 3.44FEE72 pKa = 5.57GNKK75 pKa = 10.04LRR77 pKa = 11.84VEE79 pKa = 4.49VPFNARR85 pKa = 11.84EE86 pKa = 3.83NGRR89 pKa = 11.84YY90 pKa = 7.71NARR93 pKa = 11.84GGGRR97 pKa = 11.84GMMHH101 pKa = 7.62RR102 pKa = 11.84GPKK105 pKa = 9.07SRR107 pKa = 11.84RR108 pKa = 11.84GRR110 pKa = 11.84YY111 pKa = 7.77VVEE114 pKa = 4.12VTGLPISGSWQDD126 pKa = 3.96LKK128 pKa = 11.61DD129 pKa = 3.59HH130 pKa = 6.28LRR132 pKa = 11.84EE133 pKa = 4.28AGEE136 pKa = 4.55CGHH139 pKa = 7.27ADD141 pKa = 3.39VFKK144 pKa = 10.97DD145 pKa = 3.4GTGEE149 pKa = 3.83VSFFNKK155 pKa = 10.15DD156 pKa = 3.38DD157 pKa = 3.63MLEE160 pKa = 4.66AIDD163 pKa = 4.88KK164 pKa = 10.54FNGSIFRR171 pKa = 11.84SHH173 pKa = 6.72EE174 pKa = 4.13GEE176 pKa = 3.77KK177 pKa = 10.71SKK179 pKa = 10.98ISIRR183 pKa = 11.84QKK185 pKa = 8.21KK186 pKa = 5.84TLWRR190 pKa = 11.84RR191 pKa = 11.84DD192 pKa = 3.2DD193 pKa = 3.74GGYY196 pKa = 10.49GRR198 pKa = 11.84YY199 pKa = 9.35NSRR202 pKa = 11.84HH203 pKa = 5.55RR204 pKa = 11.84SYY206 pKa = 11.42SSRR209 pKa = 11.84SYY211 pKa = 10.69SRR213 pKa = 11.84SGKK216 pKa = 9.73RR217 pKa = 11.84SYY219 pKa = 9.89TRR221 pKa = 11.84SRR223 pKa = 11.84SRR225 pKa = 11.84SYY227 pKa = 11.08SSRR230 pKa = 11.84SYY232 pKa = 10.75SRR234 pKa = 11.84SRR236 pKa = 11.84SSSSRR241 pKa = 11.84SRR243 pKa = 11.84SRR245 pKa = 11.84SMDD248 pKa = 3.04RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84DD252 pKa = 3.43TYY254 pKa = 11.14DD255 pKa = 2.76KK256 pKa = 10.51KK257 pKa = 10.76RR258 pKa = 11.84SYY260 pKa = 10.91SRR262 pKa = 11.84SLSYY266 pKa = 9.54QEE268 pKa = 3.97RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84NNRR274 pKa = 11.84TISKK278 pKa = 9.6SRR280 pKa = 11.84SRR282 pKa = 11.84SRR284 pKa = 11.84TNSRR288 pKa = 11.84SSSWSHH294 pKa = 5.94KK295 pKa = 9.54RR296 pKa = 11.84RR297 pKa = 11.84HH298 pKa = 5.26
MM1 pKa = 7.61VIRR4 pKa = 11.84EE5 pKa = 4.44SVSRR9 pKa = 11.84IYY11 pKa = 10.84VGNLPSHH18 pKa = 6.2VSSRR22 pKa = 11.84DD23 pKa = 3.23VEE25 pKa = 4.32NEE27 pKa = 3.08FRR29 pKa = 11.84KK30 pKa = 10.51YY31 pKa = 11.3GNILKK36 pKa = 10.22CDD38 pKa = 3.51VKK40 pKa = 10.59KK41 pKa = 9.2TVSGAAFAFIEE52 pKa = 4.59FEE54 pKa = 4.33DD55 pKa = 4.29ARR57 pKa = 11.84DD58 pKa = 3.3AADD61 pKa = 4.76AIKK64 pKa = 10.62EE65 pKa = 3.83KK66 pKa = 10.72DD67 pKa = 3.39GCDD70 pKa = 3.44FEE72 pKa = 5.57GNKK75 pKa = 10.04LRR77 pKa = 11.84VEE79 pKa = 4.49VPFNARR85 pKa = 11.84EE86 pKa = 3.83NGRR89 pKa = 11.84YY90 pKa = 7.71NARR93 pKa = 11.84GGGRR97 pKa = 11.84GMMHH101 pKa = 7.62RR102 pKa = 11.84GPKK105 pKa = 9.07SRR107 pKa = 11.84RR108 pKa = 11.84GRR110 pKa = 11.84YY111 pKa = 7.77VVEE114 pKa = 4.12VTGLPISGSWQDD126 pKa = 3.96LKK128 pKa = 11.61DD129 pKa = 3.59HH130 pKa = 6.28LRR132 pKa = 11.84EE133 pKa = 4.28AGEE136 pKa = 4.55CGHH139 pKa = 7.27ADD141 pKa = 3.39VFKK144 pKa = 10.97DD145 pKa = 3.4GTGEE149 pKa = 3.83VSFFNKK155 pKa = 10.15DD156 pKa = 3.38DD157 pKa = 3.63MLEE160 pKa = 4.66AIDD163 pKa = 4.88KK164 pKa = 10.54FNGSIFRR171 pKa = 11.84SHH173 pKa = 6.72EE174 pKa = 4.13GEE176 pKa = 3.77KK177 pKa = 10.71SKK179 pKa = 10.98ISIRR183 pKa = 11.84QKK185 pKa = 8.21KK186 pKa = 5.84TLWRR190 pKa = 11.84RR191 pKa = 11.84DD192 pKa = 3.2DD193 pKa = 3.74GGYY196 pKa = 10.49GRR198 pKa = 11.84YY199 pKa = 9.35NSRR202 pKa = 11.84HH203 pKa = 5.55RR204 pKa = 11.84SYY206 pKa = 11.42SSRR209 pKa = 11.84SYY211 pKa = 10.69SRR213 pKa = 11.84SGKK216 pKa = 9.73RR217 pKa = 11.84SYY219 pKa = 9.89TRR221 pKa = 11.84SRR223 pKa = 11.84SRR225 pKa = 11.84SYY227 pKa = 11.08SSRR230 pKa = 11.84SYY232 pKa = 10.75SRR234 pKa = 11.84SRR236 pKa = 11.84SSSSRR241 pKa = 11.84SRR243 pKa = 11.84SRR245 pKa = 11.84SMDD248 pKa = 3.04RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84DD252 pKa = 3.43TYY254 pKa = 11.14DD255 pKa = 2.76KK256 pKa = 10.51KK257 pKa = 10.76RR258 pKa = 11.84SYY260 pKa = 10.91SRR262 pKa = 11.84SLSYY266 pKa = 9.54QEE268 pKa = 3.97RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84NNRR274 pKa = 11.84TISKK278 pKa = 9.6SRR280 pKa = 11.84SRR282 pKa = 11.84SRR284 pKa = 11.84TNSRR288 pKa = 11.84SSSWSHH294 pKa = 5.94KK295 pKa = 9.54RR296 pKa = 11.84RR297 pKa = 11.84HH298 pKa = 5.26
Molecular weight: 34.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3932225 |
10 |
10148 |
624.1 |
73.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.966 ± 0.022 | 1.828 ± 0.016 |
6.304 ± 0.03 | 6.936 ± 0.043 |
4.472 ± 0.028 | 2.822 ± 0.027 |
2.407 ± 0.015 | 9.336 ± 0.044 |
11.767 ± 0.038 | 7.672 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.013 | 14.319 ± 0.085 |
1.973 ± 0.019 | 2.755 ± 0.015 |
2.675 ± 0.018 | 6.389 ± 0.024 |
4.109 ± 0.019 | 3.751 ± 0.02 |
0.518 ± 0.009 | 5.789 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
