
Candidatus Moduliflexus flocculans
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Candidatus Moduliflexus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
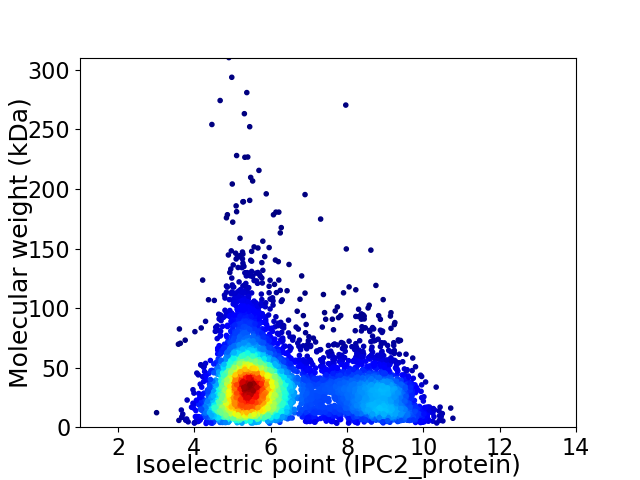
Virtual 2D-PAGE plot for 5960 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081BLX2|A0A081BLX2_9BACT Uncharacterized protein OS=Candidatus Moduliflexus flocculans OX=1499966 GN=U14_02632 PE=4 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.58HH4 pKa = 7.71YY5 pKa = 10.84MMFCVLLGVIHH16 pKa = 7.07LLSACGTGTKK26 pKa = 10.03RR27 pKa = 11.84SGALYY32 pKa = 9.8PPGANPGQLQCGGVEE47 pKa = 3.99PGKK50 pKa = 9.39TGTLVADD57 pKa = 3.52TGFRR61 pKa = 11.84PRR63 pKa = 11.84PNGFSFANYY72 pKa = 9.03GGGYY76 pKa = 9.46RR77 pKa = 11.84EE78 pKa = 4.66ANLNPEE84 pKa = 4.43AAWKK88 pKa = 10.44IFGEE92 pKa = 4.64TACKK96 pKa = 10.15RR97 pKa = 11.84MKK99 pKa = 10.42QEE101 pKa = 3.83EE102 pKa = 4.75CVPTPALKK110 pKa = 10.58LWVKK114 pKa = 9.24TMNEE118 pKa = 3.68AMGQGHH124 pKa = 6.73CEE126 pKa = 3.86GMATLSAWMFLAQDD140 pKa = 3.43KK141 pKa = 10.13LAAYY145 pKa = 7.61GKK147 pKa = 7.91PTAFDD152 pKa = 4.8LDD154 pKa = 3.79TTQADD159 pKa = 4.33LLQDD163 pKa = 3.04IAFYY167 pKa = 8.02WTLQTLEE174 pKa = 4.28PVVSEE179 pKa = 4.74FNAFQQQPPSEE190 pKa = 4.14ILNTLISTMQQKK202 pKa = 8.82TDD204 pKa = 2.84WYY206 pKa = 8.4TLGIYY211 pKa = 10.6GSVGGHH217 pKa = 6.12AVTPYY222 pKa = 10.62AVEE225 pKa = 4.02DD226 pKa = 3.58VGNGVFWIHH235 pKa = 6.55VYY237 pKa = 10.96DD238 pKa = 3.82NNYY241 pKa = 8.01PCAYY245 pKa = 9.44KK246 pKa = 10.65YY247 pKa = 11.33VEE249 pKa = 4.04VDD251 pKa = 3.25TNTEE255 pKa = 3.52TWHH258 pKa = 5.84YY259 pKa = 10.51AGAAINPAEE268 pKa = 4.9DD269 pKa = 3.41ASPWSGTTGTMDD281 pKa = 3.7LTSLSVRR288 pKa = 11.84QQSMEE293 pKa = 4.15CPFCSEE299 pKa = 4.81AQRR302 pKa = 11.84CLQNAGLSVLVNGRR316 pKa = 11.84GANIRR321 pKa = 11.84ATNAEE326 pKa = 4.16GKK328 pKa = 9.57QLSLQNGQAQNEE340 pKa = 4.32IPGAKK345 pKa = 9.43LLKK348 pKa = 10.41LKK350 pKa = 10.97GIFGEE355 pKa = 4.66DD356 pKa = 3.1RR357 pKa = 11.84ASLLILPPNAKK368 pKa = 9.9YY369 pKa = 10.67ALDD372 pKa = 3.99LGGIADD378 pKa = 4.16VEE380 pKa = 4.43EE381 pKa = 4.8KK382 pKa = 10.43SASIAMFTSCEE393 pKa = 4.11GYY395 pKa = 10.67ALSGMSLSSGQINQIMMTDD414 pKa = 3.42KK415 pKa = 11.24GRR417 pKa = 11.84FSYY420 pKa = 10.39QAGGAQSPTFEE431 pKa = 4.36AAFHH435 pKa = 6.44NPDD438 pKa = 3.82GNDD441 pKa = 3.05TFYY444 pKa = 11.07SISDD448 pKa = 3.38FDD450 pKa = 3.84IGDD453 pKa = 3.51GRR455 pKa = 11.84SISFEE460 pKa = 3.76QDD462 pKa = 2.19EE463 pKa = 4.83DD464 pKa = 3.47TGEE467 pKa = 4.62IYY469 pKa = 10.39IQDD472 pKa = 5.0DD473 pKa = 4.03DD474 pKa = 5.37PEE476 pKa = 4.77MEE478 pKa = 5.79DD479 pKa = 3.47FDD481 pKa = 6.69LDD483 pKa = 3.21IFTIDD488 pKa = 3.22TEE490 pKa = 4.88GEE492 pKa = 4.28VEE494 pKa = 4.41GISYY498 pKa = 10.94KK499 pKa = 11.03NIDD502 pKa = 3.81VEE504 pKa = 5.78DD505 pKa = 3.85EE506 pKa = 4.38GQVILHH512 pKa = 6.7FGDD515 pKa = 4.57DD516 pKa = 3.91EE517 pKa = 5.04SFEE520 pKa = 4.98LYY522 pKa = 9.92IDD524 pKa = 3.77SDD526 pKa = 4.08GDD528 pKa = 4.23GISDD532 pKa = 3.64SPPYY536 pKa = 9.9EE537 pKa = 4.34KK538 pKa = 10.92SNEE541 pKa = 4.07SPDD544 pKa = 3.25WDD546 pKa = 3.51NYY548 pKa = 11.15GYY550 pKa = 11.11DD551 pKa = 3.8DD552 pKa = 5.32ADD554 pKa = 3.56VSEE557 pKa = 4.69SSGNSFDD564 pKa = 5.82NNDD567 pKa = 3.93ADD569 pKa = 5.36DD570 pKa = 4.58SDD572 pKa = 5.36SEE574 pKa = 4.5TTYY577 pKa = 11.12SVNYY581 pKa = 8.82APSYY585 pKa = 8.5EE586 pKa = 4.31SEE588 pKa = 4.12WVDD591 pKa = 4.26DD592 pKa = 4.14YY593 pKa = 12.03ASEE596 pKa = 4.9AISEE600 pKa = 4.37KK601 pKa = 10.91YY602 pKa = 9.97EE603 pKa = 4.19EE604 pKa = 4.51NDD606 pKa = 3.7SDD608 pKa = 5.31HH609 pKa = 7.42SDD611 pKa = 4.82DD612 pKa = 5.86GDD614 pKa = 3.49WDD616 pKa = 4.25NNDD619 pKa = 4.81DD620 pKa = 5.94DD621 pKa = 6.22DD622 pKa = 7.28DD623 pKa = 5.79SDD625 pKa = 5.99HH626 pKa = 7.61GDD628 pKa = 3.64LDD630 pKa = 3.92NDD632 pKa = 3.67NGSDD636 pKa = 3.92YY637 pKa = 11.46DD638 pKa = 4.41DD639 pKa = 5.45GSDD642 pKa = 3.74TGSDD646 pKa = 3.75DD647 pKa = 4.57SGSDD651 pKa = 4.26DD652 pKa = 4.38SANDD656 pKa = 3.95PDD658 pKa = 4.74SSGSDD663 pKa = 3.16SSDD666 pKa = 3.31GSDD669 pKa = 3.23SGYY672 pKa = 11.54
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.58HH4 pKa = 7.71YY5 pKa = 10.84MMFCVLLGVIHH16 pKa = 7.07LLSACGTGTKK26 pKa = 10.03RR27 pKa = 11.84SGALYY32 pKa = 9.8PPGANPGQLQCGGVEE47 pKa = 3.99PGKK50 pKa = 9.39TGTLVADD57 pKa = 3.52TGFRR61 pKa = 11.84PRR63 pKa = 11.84PNGFSFANYY72 pKa = 9.03GGGYY76 pKa = 9.46RR77 pKa = 11.84EE78 pKa = 4.66ANLNPEE84 pKa = 4.43AAWKK88 pKa = 10.44IFGEE92 pKa = 4.64TACKK96 pKa = 10.15RR97 pKa = 11.84MKK99 pKa = 10.42QEE101 pKa = 3.83EE102 pKa = 4.75CVPTPALKK110 pKa = 10.58LWVKK114 pKa = 9.24TMNEE118 pKa = 3.68AMGQGHH124 pKa = 6.73CEE126 pKa = 3.86GMATLSAWMFLAQDD140 pKa = 3.43KK141 pKa = 10.13LAAYY145 pKa = 7.61GKK147 pKa = 7.91PTAFDD152 pKa = 4.8LDD154 pKa = 3.79TTQADD159 pKa = 4.33LLQDD163 pKa = 3.04IAFYY167 pKa = 8.02WTLQTLEE174 pKa = 4.28PVVSEE179 pKa = 4.74FNAFQQQPPSEE190 pKa = 4.14ILNTLISTMQQKK202 pKa = 8.82TDD204 pKa = 2.84WYY206 pKa = 8.4TLGIYY211 pKa = 10.6GSVGGHH217 pKa = 6.12AVTPYY222 pKa = 10.62AVEE225 pKa = 4.02DD226 pKa = 3.58VGNGVFWIHH235 pKa = 6.55VYY237 pKa = 10.96DD238 pKa = 3.82NNYY241 pKa = 8.01PCAYY245 pKa = 9.44KK246 pKa = 10.65YY247 pKa = 11.33VEE249 pKa = 4.04VDD251 pKa = 3.25TNTEE255 pKa = 3.52TWHH258 pKa = 5.84YY259 pKa = 10.51AGAAINPAEE268 pKa = 4.9DD269 pKa = 3.41ASPWSGTTGTMDD281 pKa = 3.7LTSLSVRR288 pKa = 11.84QQSMEE293 pKa = 4.15CPFCSEE299 pKa = 4.81AQRR302 pKa = 11.84CLQNAGLSVLVNGRR316 pKa = 11.84GANIRR321 pKa = 11.84ATNAEE326 pKa = 4.16GKK328 pKa = 9.57QLSLQNGQAQNEE340 pKa = 4.32IPGAKK345 pKa = 9.43LLKK348 pKa = 10.41LKK350 pKa = 10.97GIFGEE355 pKa = 4.66DD356 pKa = 3.1RR357 pKa = 11.84ASLLILPPNAKK368 pKa = 9.9YY369 pKa = 10.67ALDD372 pKa = 3.99LGGIADD378 pKa = 4.16VEE380 pKa = 4.43EE381 pKa = 4.8KK382 pKa = 10.43SASIAMFTSCEE393 pKa = 4.11GYY395 pKa = 10.67ALSGMSLSSGQINQIMMTDD414 pKa = 3.42KK415 pKa = 11.24GRR417 pKa = 11.84FSYY420 pKa = 10.39QAGGAQSPTFEE431 pKa = 4.36AAFHH435 pKa = 6.44NPDD438 pKa = 3.82GNDD441 pKa = 3.05TFYY444 pKa = 11.07SISDD448 pKa = 3.38FDD450 pKa = 3.84IGDD453 pKa = 3.51GRR455 pKa = 11.84SISFEE460 pKa = 3.76QDD462 pKa = 2.19EE463 pKa = 4.83DD464 pKa = 3.47TGEE467 pKa = 4.62IYY469 pKa = 10.39IQDD472 pKa = 5.0DD473 pKa = 4.03DD474 pKa = 5.37PEE476 pKa = 4.77MEE478 pKa = 5.79DD479 pKa = 3.47FDD481 pKa = 6.69LDD483 pKa = 3.21IFTIDD488 pKa = 3.22TEE490 pKa = 4.88GEE492 pKa = 4.28VEE494 pKa = 4.41GISYY498 pKa = 10.94KK499 pKa = 11.03NIDD502 pKa = 3.81VEE504 pKa = 5.78DD505 pKa = 3.85EE506 pKa = 4.38GQVILHH512 pKa = 6.7FGDD515 pKa = 4.57DD516 pKa = 3.91EE517 pKa = 5.04SFEE520 pKa = 4.98LYY522 pKa = 9.92IDD524 pKa = 3.77SDD526 pKa = 4.08GDD528 pKa = 4.23GISDD532 pKa = 3.64SPPYY536 pKa = 9.9EE537 pKa = 4.34KK538 pKa = 10.92SNEE541 pKa = 4.07SPDD544 pKa = 3.25WDD546 pKa = 3.51NYY548 pKa = 11.15GYY550 pKa = 11.11DD551 pKa = 3.8DD552 pKa = 5.32ADD554 pKa = 3.56VSEE557 pKa = 4.69SSGNSFDD564 pKa = 5.82NNDD567 pKa = 3.93ADD569 pKa = 5.36DD570 pKa = 4.58SDD572 pKa = 5.36SEE574 pKa = 4.5TTYY577 pKa = 11.12SVNYY581 pKa = 8.82APSYY585 pKa = 8.5EE586 pKa = 4.31SEE588 pKa = 4.12WVDD591 pKa = 4.26DD592 pKa = 4.14YY593 pKa = 12.03ASEE596 pKa = 4.9AISEE600 pKa = 4.37KK601 pKa = 10.91YY602 pKa = 9.97EE603 pKa = 4.19EE604 pKa = 4.51NDD606 pKa = 3.7SDD608 pKa = 5.31HH609 pKa = 7.42SDD611 pKa = 4.82DD612 pKa = 5.86GDD614 pKa = 3.49WDD616 pKa = 4.25NNDD619 pKa = 4.81DD620 pKa = 5.94DD621 pKa = 6.22DD622 pKa = 7.28DD623 pKa = 5.79SDD625 pKa = 5.99HH626 pKa = 7.61GDD628 pKa = 3.64LDD630 pKa = 3.92NDD632 pKa = 3.67NGSDD636 pKa = 3.92YY637 pKa = 11.46DD638 pKa = 4.41DD639 pKa = 5.45GSDD642 pKa = 3.74TGSDD646 pKa = 3.75DD647 pKa = 4.57SGSDD651 pKa = 4.26DD652 pKa = 4.38SANDD656 pKa = 3.95PDD658 pKa = 4.74SSGSDD663 pKa = 3.16SSDD666 pKa = 3.31GSDD669 pKa = 3.23SGYY672 pKa = 11.54
Molecular weight: 73.07 kDa
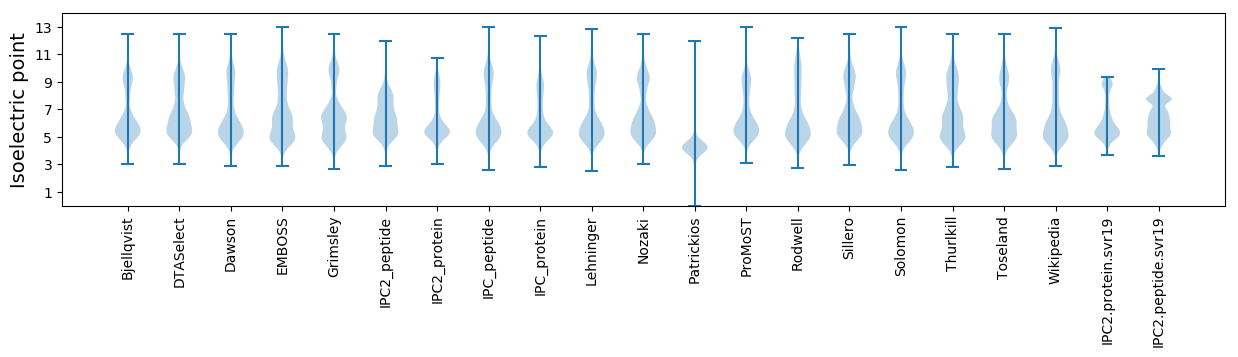
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081BQX6|A0A081BQX6_9BACT Putative ABC transporter permease protein OS=Candidatus Moduliflexus flocculans OX=1499966 GN=U14_05081 PE=4 SV=1
MM1 pKa = 7.37GRR3 pKa = 11.84LFHH6 pKa = 6.36PRR8 pKa = 11.84QRR10 pKa = 11.84EE11 pKa = 3.9LEE13 pKa = 4.18LPQRR17 pKa = 11.84QLLRR21 pKa = 11.84ARR23 pKa = 11.84RR24 pKa = 11.84SFLTMMWIIWSFDD37 pKa = 3.48PLGGLGGDD45 pKa = 3.79SPPAKK50 pKa = 10.19KK51 pKa = 10.29FFLGTTGIRR60 pKa = 11.84GRR62 pKa = 11.84KK63 pKa = 8.73DD64 pKa = 2.79RR65 pKa = 11.84GIILAYY71 pKa = 9.08TYY73 pKa = 9.38PCRR76 pKa = 11.84SFKK79 pKa = 11.01VCLTDD84 pKa = 3.67SIGRR88 pKa = 11.84GEE90 pKa = 4.54PPCSPEE96 pKa = 3.46PHH98 pKa = 6.22ARR100 pKa = 11.84CPNHH104 pKa = 6.26TRR106 pKa = 11.84DD107 pKa = 3.37ARR109 pKa = 11.84TTRR112 pKa = 11.84EE113 pKa = 3.78MPEE116 pKa = 3.57QQGRR120 pKa = 11.84HH121 pKa = 5.46GGLPLPAVVCRR132 pKa = 11.84KK133 pKa = 10.01NPLTLLHH140 pKa = 5.97TRR142 pKa = 11.84RR143 pKa = 11.84KK144 pKa = 10.29LSDD147 pKa = 3.27TFKK150 pKa = 11.08VSDD153 pKa = 3.71SSAHH157 pKa = 6.24NVLCQYY163 pKa = 11.15RR164 pKa = 11.84PMRR167 pKa = 11.84SSIAVFIASRR177 pKa = 11.84VRR179 pKa = 11.84AKK181 pKa = 10.69SSSEE185 pKa = 3.88CANEE189 pKa = 3.6TNAASNCDD197 pKa = 3.36GARR200 pKa = 3.31
MM1 pKa = 7.37GRR3 pKa = 11.84LFHH6 pKa = 6.36PRR8 pKa = 11.84QRR10 pKa = 11.84EE11 pKa = 3.9LEE13 pKa = 4.18LPQRR17 pKa = 11.84QLLRR21 pKa = 11.84ARR23 pKa = 11.84RR24 pKa = 11.84SFLTMMWIIWSFDD37 pKa = 3.48PLGGLGGDD45 pKa = 3.79SPPAKK50 pKa = 10.19KK51 pKa = 10.29FFLGTTGIRR60 pKa = 11.84GRR62 pKa = 11.84KK63 pKa = 8.73DD64 pKa = 2.79RR65 pKa = 11.84GIILAYY71 pKa = 9.08TYY73 pKa = 9.38PCRR76 pKa = 11.84SFKK79 pKa = 11.01VCLTDD84 pKa = 3.67SIGRR88 pKa = 11.84GEE90 pKa = 4.54PPCSPEE96 pKa = 3.46PHH98 pKa = 6.22ARR100 pKa = 11.84CPNHH104 pKa = 6.26TRR106 pKa = 11.84DD107 pKa = 3.37ARR109 pKa = 11.84TTRR112 pKa = 11.84EE113 pKa = 3.78MPEE116 pKa = 3.57QQGRR120 pKa = 11.84HH121 pKa = 5.46GGLPLPAVVCRR132 pKa = 11.84KK133 pKa = 10.01NPLTLLHH140 pKa = 5.97TRR142 pKa = 11.84RR143 pKa = 11.84KK144 pKa = 10.29LSDD147 pKa = 3.27TFKK150 pKa = 11.08VSDD153 pKa = 3.71SSAHH157 pKa = 6.24NVLCQYY163 pKa = 11.15RR164 pKa = 11.84PMRR167 pKa = 11.84SSIAVFIASRR177 pKa = 11.84VRR179 pKa = 11.84AKK181 pKa = 10.69SSSEE185 pKa = 3.88CANEE189 pKa = 3.6TNAASNCDD197 pKa = 3.36GARR200 pKa = 3.31
Molecular weight: 22.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2000297 |
29 |
2846 |
335.6 |
37.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.121 ± 0.036 | 1.101 ± 0.012 |
5.084 ± 0.024 | 6.666 ± 0.033 |
4.556 ± 0.025 | 6.496 ± 0.025 |
2.316 ± 0.016 | 6.772 ± 0.026 |
4.693 ± 0.025 | 10.352 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.62 ± 0.014 | 3.547 ± 0.019 |
4.372 ± 0.023 | 4.674 ± 0.024 |
5.701 ± 0.025 | 5.508 ± 0.02 |
5.446 ± 0.019 | 6.529 ± 0.025 |
1.218 ± 0.014 | 3.226 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
