
Chlorobium phaeobacteroides (strain DSM 266)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae; Chlorobium/Pelodictyon group; Chlorobium; Chlorobium phaeobacteroides
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
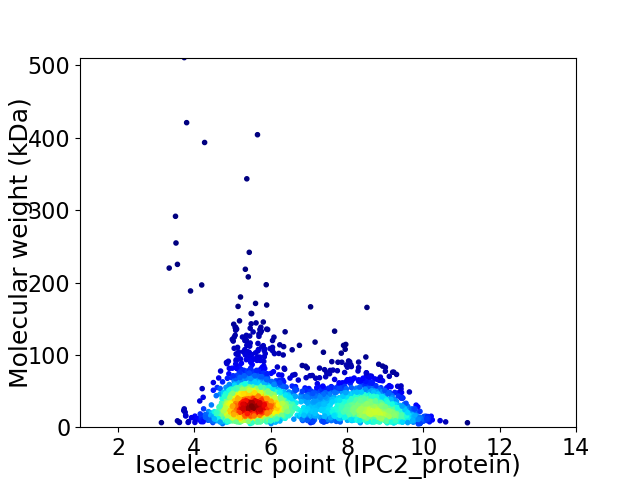
Virtual 2D-PAGE plot for 2579 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A1BDM2|A1BDM2_CHLPD Molybdenum ABC transporter periplasmic molybdate-binding protein OS=Chlorobium phaeobacteroides (strain DSM 266) OX=290317 GN=Cpha266_0441 PE=3 SV=1
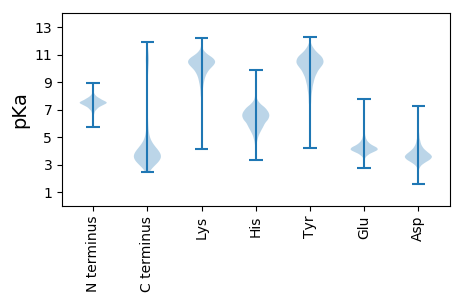
MM1 pKa = 7.42KK2 pKa = 10.44KK3 pKa = 10.14SLSLLAVLAVVGFASTPVQAADD25 pKa = 3.95HH26 pKa = 6.15YY27 pKa = 8.93VSGMAGISWMQDD39 pKa = 2.66SSIDD43 pKa = 3.84LPLNLNEE50 pKa = 4.12VFGVGIDD57 pKa = 3.67LGYY60 pKa = 10.91SSGFTAVGAVGCDD73 pKa = 3.42YY74 pKa = 11.44GSTRR78 pKa = 11.84LEE80 pKa = 4.18AEE82 pKa = 4.01VGYY85 pKa = 7.84QTNDD89 pKa = 2.6IDD91 pKa = 5.4SITGSFDD98 pKa = 3.15GDD100 pKa = 3.97SASLALSGNAKK111 pKa = 9.97VLSLMANGFYY121 pKa = 10.8DD122 pKa = 3.85FDD124 pKa = 5.26LGGVEE129 pKa = 4.9LYY131 pKa = 11.3AMAGIGVAQVSINDD145 pKa = 3.56LTLADD150 pKa = 4.31TNDD153 pKa = 3.79LGISADD159 pKa = 4.97DD160 pKa = 3.46IDD162 pKa = 5.27DD163 pKa = 4.13ANEE166 pKa = 3.83YY167 pKa = 10.53LSRR170 pKa = 11.84RR171 pKa = 11.84LGAPVDD177 pKa = 4.42LNALGLNVNEE187 pKa = 4.18TTLAYY192 pKa = 9.93QVGAGLAVPIGDD204 pKa = 5.1GIMLDD209 pKa = 2.89ARR211 pKa = 11.84YY212 pKa = 9.76RR213 pKa = 11.84YY214 pKa = 8.81FATTDD219 pKa = 3.5FTMPLDD225 pKa = 4.15LNTNISSHH233 pKa = 5.66SALLGLRR240 pKa = 11.84VNLL243 pKa = 4.21
MM1 pKa = 7.42KK2 pKa = 10.44KK3 pKa = 10.14SLSLLAVLAVVGFASTPVQAADD25 pKa = 3.95HH26 pKa = 6.15YY27 pKa = 8.93VSGMAGISWMQDD39 pKa = 2.66SSIDD43 pKa = 3.84LPLNLNEE50 pKa = 4.12VFGVGIDD57 pKa = 3.67LGYY60 pKa = 10.91SSGFTAVGAVGCDD73 pKa = 3.42YY74 pKa = 11.44GSTRR78 pKa = 11.84LEE80 pKa = 4.18AEE82 pKa = 4.01VGYY85 pKa = 7.84QTNDD89 pKa = 2.6IDD91 pKa = 5.4SITGSFDD98 pKa = 3.15GDD100 pKa = 3.97SASLALSGNAKK111 pKa = 9.97VLSLMANGFYY121 pKa = 10.8DD122 pKa = 3.85FDD124 pKa = 5.26LGGVEE129 pKa = 4.9LYY131 pKa = 11.3AMAGIGVAQVSINDD145 pKa = 3.56LTLADD150 pKa = 4.31TNDD153 pKa = 3.79LGISADD159 pKa = 4.97DD160 pKa = 3.46IDD162 pKa = 5.27DD163 pKa = 4.13ANEE166 pKa = 3.83YY167 pKa = 10.53LSRR170 pKa = 11.84RR171 pKa = 11.84LGAPVDD177 pKa = 4.42LNALGLNVNEE187 pKa = 4.18TTLAYY192 pKa = 9.93QVGAGLAVPIGDD204 pKa = 5.1GIMLDD209 pKa = 2.89ARR211 pKa = 11.84YY212 pKa = 9.76RR213 pKa = 11.84YY214 pKa = 8.81FATTDD219 pKa = 3.5FTMPLDD225 pKa = 4.15LNTNISSHH233 pKa = 5.66SALLGLRR240 pKa = 11.84VNLL243 pKa = 4.21
Molecular weight: 25.33 kDa
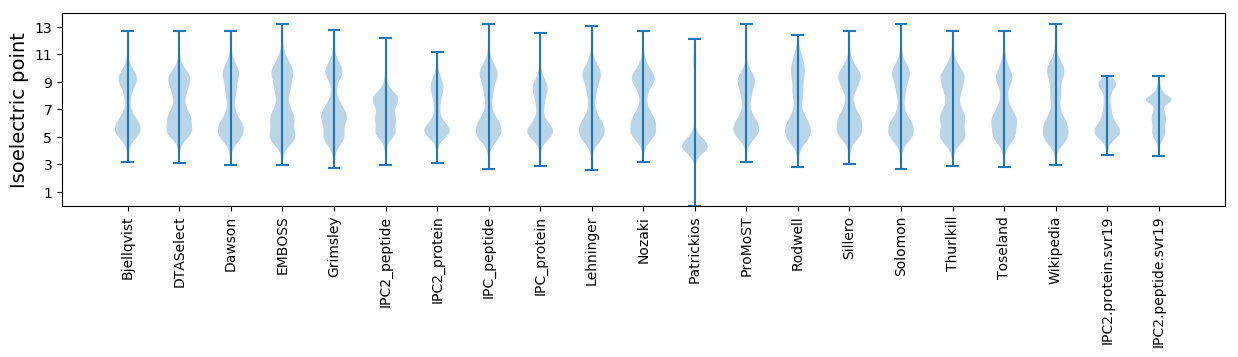
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A1BCI7|A1BCI7_CHLPD Geranylgeranyl reductase OS=Chlorobium phaeobacteroides (strain DSM 266) OX=290317 GN=Cpha266_0044 PE=3 SV=1
MM1 pKa = 7.33KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.66QPRR8 pKa = 11.84NRR10 pKa = 11.84KK11 pKa = 8.8RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.88HH16 pKa = 4.47GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTKK25 pKa = 9.43NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84HH40 pKa = 4.91SLSVSSAMGTAGQQ53 pKa = 3.64
MM1 pKa = 7.33KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.66QPRR8 pKa = 11.84NRR10 pKa = 11.84KK11 pKa = 8.8RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.88HH16 pKa = 4.47GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTKK25 pKa = 9.43NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84HH40 pKa = 4.91SLSVSSAMGTAGQQ53 pKa = 3.64
Molecular weight: 6.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
851888 |
38 |
4876 |
330.3 |
36.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.298 ± 0.056 | 1.142 ± 0.024 |
5.379 ± 0.063 | 6.628 ± 0.051 |
4.452 ± 0.041 | 7.379 ± 0.136 |
2.118 ± 0.026 | 6.634 ± 0.041 |
5.471 ± 0.058 | 10.198 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.517 ± 0.029 | 3.83 ± 0.049 |
4.126 ± 0.041 | 3.289 ± 0.031 |
5.693 ± 0.055 | 6.804 ± 0.058 |
5.201 ± 0.065 | 6.691 ± 0.044 |
1.061 ± 0.019 | 3.088 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
