
Planomonospora sphaerica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiaceae;
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
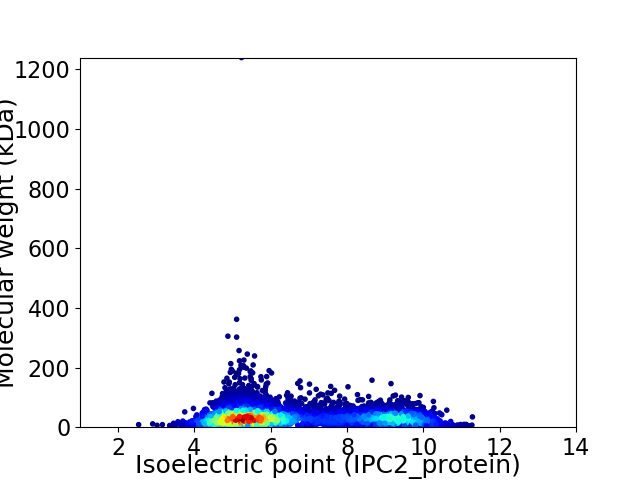
Virtual 2D-PAGE plot for 7159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A171DKX2|A0A171DKX2_9ACTN Flagellar basal body rod protein FlgB OS=Planomonospora sphaerica OX=161355 GN=PS9374_05152 PE=3 SV=1
MM1 pKa = 7.7PAGSPLRR8 pKa = 11.84LPFLRR13 pKa = 11.84SGPARR18 pKa = 11.84RR19 pKa = 11.84AAAADD24 pKa = 3.66APAEE28 pKa = 4.16SLALATTTLAYY39 pKa = 9.99VVNGGDD45 pKa = 3.62GTVSVIDD52 pKa = 3.66TTTNTVTATITVGNDD67 pKa = 3.24PYY69 pKa = 10.75RR70 pKa = 11.84VAVTPDD76 pKa = 2.87GTTAYY81 pKa = 10.33VPNADD86 pKa = 3.76GGTVSVIDD94 pKa = 3.69TTTNTVTATITVGNSPYY111 pKa = 10.57GVAVTPDD118 pKa = 3.14GTTAYY123 pKa = 10.3VPNSDD128 pKa = 3.67GGTVSVIDD136 pKa = 3.69TTTNTVTATITVGNDD151 pKa = 3.38PLGVAVSPDD160 pKa = 3.05GATAYY165 pKa = 10.11VVNQSDD171 pKa = 3.91DD172 pKa = 3.77TVSVIDD178 pKa = 3.83TATGTVTATIDD189 pKa = 3.26VDD191 pKa = 3.67DD192 pKa = 5.47RR193 pKa = 11.84PISVAFGPDD202 pKa = 2.79GATAYY207 pKa = 9.88VANPGDD213 pKa = 3.85GTVSVIDD220 pKa = 3.81TATGTVTATITVGNSPYY237 pKa = 10.83AVAVTPDD244 pKa = 3.11GATAYY249 pKa = 10.73VPGNSGGTVSVIDD262 pKa = 3.81TATGTVVDD270 pKa = 5.4TITVSADD277 pKa = 3.25PYY279 pKa = 10.58DD280 pKa = 4.02AAVSPDD286 pKa = 3.1GATVYY291 pKa = 8.78VTHH294 pKa = 7.28EE295 pKa = 4.65DD296 pKa = 3.46GGTVSVIDD304 pKa = 3.81TATGTVVDD312 pKa = 5.47TITAGDD318 pKa = 4.04DD319 pKa = 3.44PRR321 pKa = 11.84GVAVAAVPVSSAVTLTASPDD341 pKa = 3.45PAQVHH346 pKa = 5.81RR347 pKa = 11.84PVTLTAQVTCTTGTPAGTVRR367 pKa = 11.84FYY369 pKa = 11.46DD370 pKa = 4.11GAALLGTRR378 pKa = 11.84PLNAAGTAVLTTSGLDD394 pKa = 3.42AGDD397 pKa = 3.98HH398 pKa = 6.17QITAVYY404 pKa = 9.63SGDD407 pKa = 4.24GICPPAVSPPLLLTVNKK424 pKa = 9.78GVSSVQVLAVPNPALVFLPVTLTARR449 pKa = 11.84VTCDD453 pKa = 2.69SDD455 pKa = 3.74IPTGTVRR462 pKa = 11.84FMDD465 pKa = 4.13GAVTLGTVPVLAGAAVLITAFRR487 pKa = 11.84SRR489 pKa = 11.84GPHH492 pKa = 6.06QITAIYY498 pKa = 10.1SGDD501 pKa = 3.74ADD503 pKa = 4.54CEE505 pKa = 4.26PSASPPRR512 pKa = 11.84TVTVIGLLL520 pKa = 3.64
MM1 pKa = 7.7PAGSPLRR8 pKa = 11.84LPFLRR13 pKa = 11.84SGPARR18 pKa = 11.84RR19 pKa = 11.84AAAADD24 pKa = 3.66APAEE28 pKa = 4.16SLALATTTLAYY39 pKa = 9.99VVNGGDD45 pKa = 3.62GTVSVIDD52 pKa = 3.66TTTNTVTATITVGNDD67 pKa = 3.24PYY69 pKa = 10.75RR70 pKa = 11.84VAVTPDD76 pKa = 2.87GTTAYY81 pKa = 10.33VPNADD86 pKa = 3.76GGTVSVIDD94 pKa = 3.69TTTNTVTATITVGNSPYY111 pKa = 10.57GVAVTPDD118 pKa = 3.14GTTAYY123 pKa = 10.3VPNSDD128 pKa = 3.67GGTVSVIDD136 pKa = 3.69TTTNTVTATITVGNDD151 pKa = 3.38PLGVAVSPDD160 pKa = 3.05GATAYY165 pKa = 10.11VVNQSDD171 pKa = 3.91DD172 pKa = 3.77TVSVIDD178 pKa = 3.83TATGTVTATIDD189 pKa = 3.26VDD191 pKa = 3.67DD192 pKa = 5.47RR193 pKa = 11.84PISVAFGPDD202 pKa = 2.79GATAYY207 pKa = 9.88VANPGDD213 pKa = 3.85GTVSVIDD220 pKa = 3.81TATGTVTATITVGNSPYY237 pKa = 10.83AVAVTPDD244 pKa = 3.11GATAYY249 pKa = 10.73VPGNSGGTVSVIDD262 pKa = 3.81TATGTVVDD270 pKa = 5.4TITVSADD277 pKa = 3.25PYY279 pKa = 10.58DD280 pKa = 4.02AAVSPDD286 pKa = 3.1GATVYY291 pKa = 8.78VTHH294 pKa = 7.28EE295 pKa = 4.65DD296 pKa = 3.46GGTVSVIDD304 pKa = 3.81TATGTVVDD312 pKa = 5.47TITAGDD318 pKa = 4.04DD319 pKa = 3.44PRR321 pKa = 11.84GVAVAAVPVSSAVTLTASPDD341 pKa = 3.45PAQVHH346 pKa = 5.81RR347 pKa = 11.84PVTLTAQVTCTTGTPAGTVRR367 pKa = 11.84FYY369 pKa = 11.46DD370 pKa = 4.11GAALLGTRR378 pKa = 11.84PLNAAGTAVLTTSGLDD394 pKa = 3.42AGDD397 pKa = 3.98HH398 pKa = 6.17QITAVYY404 pKa = 9.63SGDD407 pKa = 4.24GICPPAVSPPLLLTVNKK424 pKa = 9.78GVSSVQVLAVPNPALVFLPVTLTARR449 pKa = 11.84VTCDD453 pKa = 2.69SDD455 pKa = 3.74IPTGTVRR462 pKa = 11.84FMDD465 pKa = 4.13GAVTLGTVPVLAGAAVLITAFRR487 pKa = 11.84SRR489 pKa = 11.84GPHH492 pKa = 6.06QITAIYY498 pKa = 10.1SGDD501 pKa = 3.74ADD503 pKa = 4.54CEE505 pKa = 4.26PSASPPRR512 pKa = 11.84TVTVIGLLL520 pKa = 3.64
Molecular weight: 51.33 kDa
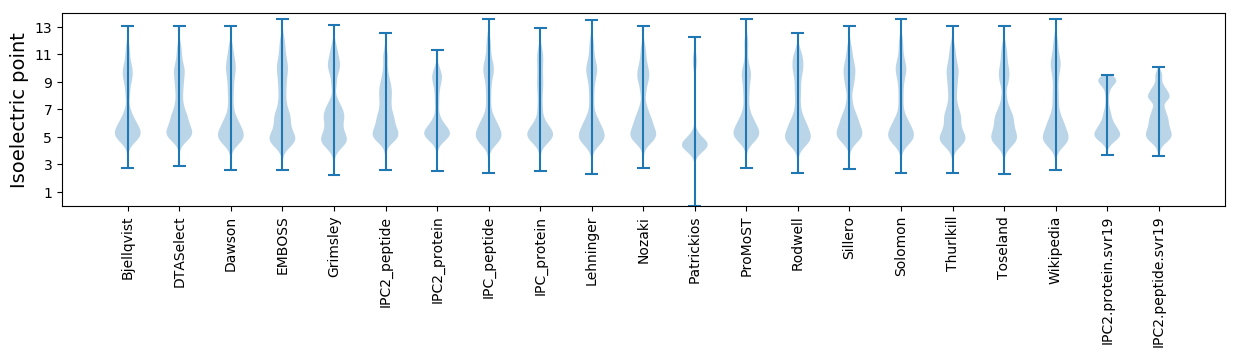
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161LL11|A0A161LL11_9ACTN ATPase OS=Planomonospora sphaerica OX=161355 GN=PS9374_05774 PE=4 SV=1
MM1 pKa = 6.82WRR3 pKa = 11.84ARR5 pKa = 11.84SSWPSSRR12 pKa = 11.84RR13 pKa = 11.84WASSFVRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84PSKK26 pKa = 10.48HH27 pKa = 5.31RR28 pKa = 11.84WSAGSRR34 pKa = 11.84KK35 pKa = 9.24LWACPRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84PTRR47 pKa = 11.84VPEE50 pKa = 4.02GRR52 pKa = 11.84PGRR55 pKa = 11.84LRR57 pKa = 11.84RR58 pKa = 11.84PPGRR62 pKa = 11.84PRR64 pKa = 11.84ARR66 pKa = 11.84PAALRR71 pKa = 11.84RR72 pKa = 11.84PPRR75 pKa = 11.84FPGRR79 pKa = 11.84DD80 pKa = 3.14RR81 pKa = 11.84LPSRR85 pKa = 11.84VRR87 pKa = 11.84GRR89 pKa = 11.84LRR91 pKa = 11.84VLRR94 pKa = 11.84PPRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84PRR101 pKa = 11.84SPRR104 pKa = 11.84FSSSPRR110 pKa = 11.84SSSSRR115 pKa = 11.84RR116 pKa = 11.84CSSSRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84SSRR126 pKa = 11.84RR127 pKa = 11.84VSRR130 pKa = 11.84RR131 pKa = 11.84LRR133 pKa = 11.84PRR135 pKa = 11.84GPRR138 pKa = 11.84APAVPVPSRR147 pKa = 11.84ARR149 pKa = 11.84ARR151 pKa = 11.84ARR153 pKa = 11.84ARR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84VPRR160 pKa = 11.84RR161 pKa = 11.84PARR164 pKa = 11.84RR165 pKa = 11.84PVPPAVPVPLRR176 pKa = 11.84ARR178 pKa = 11.84VPPPGLPVVPVPSRR192 pKa = 11.84VRR194 pKa = 11.84VRR196 pKa = 11.84ARR198 pKa = 11.84AARR201 pKa = 11.84DD202 pKa = 3.07RR203 pKa = 11.84ATTRR207 pKa = 11.84SPRR210 pKa = 11.84TPAAWARR217 pKa = 11.84PVRR220 pKa = 11.84RR221 pKa = 11.84VRR223 pKa = 11.84AAPAVPVTAVPVPAAVPVRR242 pKa = 11.84TAARR246 pKa = 11.84ATVRR250 pKa = 11.84LRR252 pKa = 11.84VPRR255 pKa = 11.84APVPARR261 pKa = 11.84PVRR264 pKa = 11.84PVPVRR269 pKa = 11.84GPVRR273 pKa = 11.84AVRR276 pKa = 11.84VRR278 pKa = 11.84AAVPARR284 pKa = 11.84PVRR287 pKa = 11.84APAAARR293 pKa = 11.84VPAVPVPEE301 pKa = 4.06VRR303 pKa = 11.84VPARR307 pKa = 3.43
MM1 pKa = 6.82WRR3 pKa = 11.84ARR5 pKa = 11.84SSWPSSRR12 pKa = 11.84RR13 pKa = 11.84WASSFVRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84PSKK26 pKa = 10.48HH27 pKa = 5.31RR28 pKa = 11.84WSAGSRR34 pKa = 11.84KK35 pKa = 9.24LWACPRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84PTRR47 pKa = 11.84VPEE50 pKa = 4.02GRR52 pKa = 11.84PGRR55 pKa = 11.84LRR57 pKa = 11.84RR58 pKa = 11.84PPGRR62 pKa = 11.84PRR64 pKa = 11.84ARR66 pKa = 11.84PAALRR71 pKa = 11.84RR72 pKa = 11.84PPRR75 pKa = 11.84FPGRR79 pKa = 11.84DD80 pKa = 3.14RR81 pKa = 11.84LPSRR85 pKa = 11.84VRR87 pKa = 11.84GRR89 pKa = 11.84LRR91 pKa = 11.84VLRR94 pKa = 11.84PPRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84PRR101 pKa = 11.84SPRR104 pKa = 11.84FSSSPRR110 pKa = 11.84SSSSRR115 pKa = 11.84RR116 pKa = 11.84CSSSRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84SSRR126 pKa = 11.84RR127 pKa = 11.84VSRR130 pKa = 11.84RR131 pKa = 11.84LRR133 pKa = 11.84PRR135 pKa = 11.84GPRR138 pKa = 11.84APAVPVPSRR147 pKa = 11.84ARR149 pKa = 11.84ARR151 pKa = 11.84ARR153 pKa = 11.84ARR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84VPRR160 pKa = 11.84RR161 pKa = 11.84PARR164 pKa = 11.84RR165 pKa = 11.84PVPPAVPVPLRR176 pKa = 11.84ARR178 pKa = 11.84VPPPGLPVVPVPSRR192 pKa = 11.84VRR194 pKa = 11.84VRR196 pKa = 11.84ARR198 pKa = 11.84AARR201 pKa = 11.84DD202 pKa = 3.07RR203 pKa = 11.84ATTRR207 pKa = 11.84SPRR210 pKa = 11.84TPAAWARR217 pKa = 11.84PVRR220 pKa = 11.84RR221 pKa = 11.84VRR223 pKa = 11.84AAPAVPVTAVPVPAAVPVRR242 pKa = 11.84TAARR246 pKa = 11.84ATVRR250 pKa = 11.84LRR252 pKa = 11.84VPRR255 pKa = 11.84APVPARR261 pKa = 11.84PVRR264 pKa = 11.84PVPVRR269 pKa = 11.84GPVRR273 pKa = 11.84AVRR276 pKa = 11.84VRR278 pKa = 11.84AAVPARR284 pKa = 11.84PVRR287 pKa = 11.84APAAARR293 pKa = 11.84VPAVPVPEE301 pKa = 4.06VRR303 pKa = 11.84VPARR307 pKa = 3.43
Molecular weight: 34.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2355000 |
29 |
11486 |
329.0 |
35.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.06 ± 0.035 | 0.775 ± 0.008 |
5.666 ± 0.024 | 5.834 ± 0.028 |
2.716 ± 0.016 | 9.867 ± 0.034 |
2.162 ± 0.015 | 3.196 ± 0.019 |
1.748 ± 0.02 | 10.497 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.797 ± 0.013 | 1.551 ± 0.015 |
6.337 ± 0.031 | 2.405 ± 0.02 |
8.697 ± 0.033 | 4.875 ± 0.019 |
5.655 ± 0.024 | 8.708 ± 0.031 |
1.502 ± 0.012 | 1.95 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
