
Aeromicrobium sp. Root344
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; unclassified Aeromicrobium
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
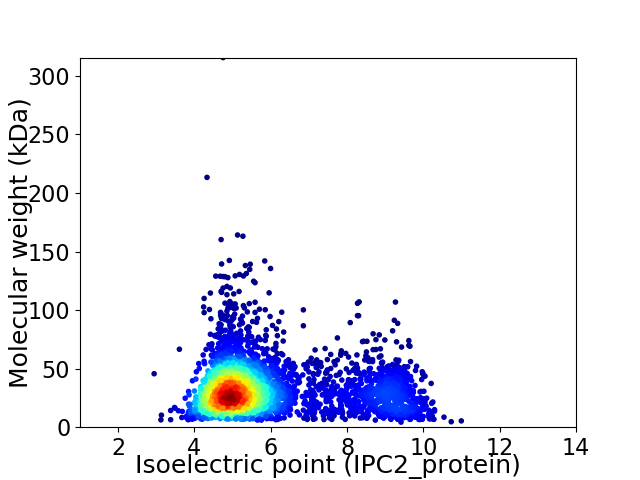
Virtual 2D-PAGE plot for 3738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6VSF3|A0A0Q6VSF3_9ACTN Uncharacterized protein OS=Aeromicrobium sp. Root344 OX=1736521 GN=ASC61_07745 PE=4 SV=1
MM1 pKa = 7.41RR2 pKa = 11.84AVRR5 pKa = 11.84GLFALLLAVILSLTFTAAHH24 pKa = 7.02ADD26 pKa = 3.79DD27 pKa = 5.59DD28 pKa = 4.93PGGGTPEE35 pKa = 4.05TSQTPEE41 pKa = 4.42PSDD44 pKa = 3.67QPTDD48 pKa = 3.62PAPTPTEE55 pKa = 4.2TTPSPSEE62 pKa = 3.82PTPTEE67 pKa = 3.98TTPTPSEE74 pKa = 4.19PKK76 pKa = 10.07PSDD79 pKa = 3.26GTPTGEE85 pKa = 4.34PGVEE89 pKa = 4.13PTDD92 pKa = 3.95QPTKK96 pKa = 9.89PADD99 pKa = 3.75RR100 pKa = 11.84PGRR103 pKa = 11.84DD104 pKa = 3.09ATVKK108 pKa = 10.49AKK110 pKa = 9.95PAIEE114 pKa = 4.21GFVAEE119 pKa = 4.71IAAALAAPVANDD131 pKa = 3.55DD132 pKa = 3.89VFSVVTNTGAFPSNIGLLRR151 pKa = 11.84PGIIGVYY158 pKa = 10.23RR159 pKa = 11.84NDD161 pKa = 3.13VPTAGGDD168 pKa = 3.34QTPNSIVLMNDD179 pKa = 2.66VDD181 pKa = 5.35HH182 pKa = 6.84GTLDD186 pKa = 3.74LNGDD190 pKa = 3.9GGFHH194 pKa = 5.9YY195 pKa = 10.65VPEE198 pKa = 4.24VGYY201 pKa = 10.81SGTDD205 pKa = 3.02TFDD208 pKa = 2.96YY209 pKa = 10.66RR210 pKa = 11.84YY211 pKa = 8.97ATDD214 pKa = 4.46DD215 pKa = 4.17GEE217 pKa = 5.33SNTATVTIVVGNGGLIATDD236 pKa = 3.49DD237 pKa = 4.38HH238 pKa = 7.11YY239 pKa = 11.79VTAPDD244 pKa = 3.18TALQVSVPGVSSNDD258 pKa = 3.25SEE260 pKa = 4.02IAYY263 pKa = 10.49LGFQVTNNVDD273 pKa = 3.46HH274 pKa = 6.72GTLTQPGFLPSGGFTYY290 pKa = 10.39TPDD293 pKa = 3.63PGFVGVDD300 pKa = 3.14TFTYY304 pKa = 9.98RR305 pKa = 11.84YY306 pKa = 9.35RR307 pKa = 11.84PLPFGPYY314 pKa = 10.09SNTATVTIIVGEE326 pKa = 4.13PPVAEE331 pKa = 4.77DD332 pKa = 3.69DD333 pKa = 4.39AYY335 pKa = 11.54SIAANSALHH344 pKa = 4.8VTAPGVRR351 pKa = 11.84DD352 pKa = 3.57NDD354 pKa = 3.9TGTGTVAVTGDD365 pKa = 3.67VSHH368 pKa = 6.45GTLDD372 pKa = 3.42LHH374 pKa = 6.8TDD376 pKa = 3.38GSFDD380 pKa = 3.81YY381 pKa = 11.12APTADD386 pKa = 4.0FVGVDD391 pKa = 2.99SFTYY395 pKa = 10.48RR396 pKa = 11.84FTDD399 pKa = 3.5PVTTLSDD406 pKa = 3.5VATVTLRR413 pKa = 11.84VTPVAADD420 pKa = 3.67DD421 pKa = 4.98AYY423 pKa = 10.05QTNAEE428 pKa = 4.11STLNVNAPGVFGNDD442 pKa = 3.21TPTSGGTPSVTDD454 pKa = 4.1DD455 pKa = 3.87VDD457 pKa = 5.77DD458 pKa = 4.44GTLTLNDD465 pKa = 4.94DD466 pKa = 3.91GSFTYY471 pKa = 10.43TPDD474 pKa = 3.12AGFAGVDD481 pKa = 3.31TFSYY485 pKa = 10.48SFQSTEE491 pKa = 3.69LDD493 pKa = 3.76SNVATVTITVSPVAVDD509 pKa = 4.49DD510 pKa = 4.25EE511 pKa = 5.26VSAQQCDD518 pKa = 3.62DD519 pKa = 3.57RR520 pKa = 11.84TFTPLANDD528 pKa = 3.84AGVDD532 pKa = 3.72PLGSVPTIIAPPEE545 pKa = 4.61HH546 pKa = 6.37GTLTYY551 pKa = 10.62PEE553 pKa = 4.89DD554 pKa = 3.67LAGGALIYY562 pKa = 11.02DD563 pKa = 4.31PDD565 pKa = 4.96DD566 pKa = 5.11DD567 pKa = 5.27FVGDD571 pKa = 3.82DD572 pKa = 3.29TFTYY576 pKa = 10.07TFATQGVVSNAATVTIHH593 pKa = 5.29VTACEE598 pKa = 4.22SGDD601 pKa = 4.0GGNSDD606 pKa = 5.53DD607 pKa = 5.19GDD609 pKa = 3.7EE610 pKa = 4.66ALPDD614 pKa = 3.73TGSPASPWLLLVAGFATFAGVVLIRR639 pKa = 11.84RR640 pKa = 11.84RR641 pKa = 11.84GGAQAA646 pKa = 4.25
MM1 pKa = 7.41RR2 pKa = 11.84AVRR5 pKa = 11.84GLFALLLAVILSLTFTAAHH24 pKa = 7.02ADD26 pKa = 3.79DD27 pKa = 5.59DD28 pKa = 4.93PGGGTPEE35 pKa = 4.05TSQTPEE41 pKa = 4.42PSDD44 pKa = 3.67QPTDD48 pKa = 3.62PAPTPTEE55 pKa = 4.2TTPSPSEE62 pKa = 3.82PTPTEE67 pKa = 3.98TTPTPSEE74 pKa = 4.19PKK76 pKa = 10.07PSDD79 pKa = 3.26GTPTGEE85 pKa = 4.34PGVEE89 pKa = 4.13PTDD92 pKa = 3.95QPTKK96 pKa = 9.89PADD99 pKa = 3.75RR100 pKa = 11.84PGRR103 pKa = 11.84DD104 pKa = 3.09ATVKK108 pKa = 10.49AKK110 pKa = 9.95PAIEE114 pKa = 4.21GFVAEE119 pKa = 4.71IAAALAAPVANDD131 pKa = 3.55DD132 pKa = 3.89VFSVVTNTGAFPSNIGLLRR151 pKa = 11.84PGIIGVYY158 pKa = 10.23RR159 pKa = 11.84NDD161 pKa = 3.13VPTAGGDD168 pKa = 3.34QTPNSIVLMNDD179 pKa = 2.66VDD181 pKa = 5.35HH182 pKa = 6.84GTLDD186 pKa = 3.74LNGDD190 pKa = 3.9GGFHH194 pKa = 5.9YY195 pKa = 10.65VPEE198 pKa = 4.24VGYY201 pKa = 10.81SGTDD205 pKa = 3.02TFDD208 pKa = 2.96YY209 pKa = 10.66RR210 pKa = 11.84YY211 pKa = 8.97ATDD214 pKa = 4.46DD215 pKa = 4.17GEE217 pKa = 5.33SNTATVTIVVGNGGLIATDD236 pKa = 3.49DD237 pKa = 4.38HH238 pKa = 7.11YY239 pKa = 11.79VTAPDD244 pKa = 3.18TALQVSVPGVSSNDD258 pKa = 3.25SEE260 pKa = 4.02IAYY263 pKa = 10.49LGFQVTNNVDD273 pKa = 3.46HH274 pKa = 6.72GTLTQPGFLPSGGFTYY290 pKa = 10.39TPDD293 pKa = 3.63PGFVGVDD300 pKa = 3.14TFTYY304 pKa = 9.98RR305 pKa = 11.84YY306 pKa = 9.35RR307 pKa = 11.84PLPFGPYY314 pKa = 10.09SNTATVTIIVGEE326 pKa = 4.13PPVAEE331 pKa = 4.77DD332 pKa = 3.69DD333 pKa = 4.39AYY335 pKa = 11.54SIAANSALHH344 pKa = 4.8VTAPGVRR351 pKa = 11.84DD352 pKa = 3.57NDD354 pKa = 3.9TGTGTVAVTGDD365 pKa = 3.67VSHH368 pKa = 6.45GTLDD372 pKa = 3.42LHH374 pKa = 6.8TDD376 pKa = 3.38GSFDD380 pKa = 3.81YY381 pKa = 11.12APTADD386 pKa = 4.0FVGVDD391 pKa = 2.99SFTYY395 pKa = 10.48RR396 pKa = 11.84FTDD399 pKa = 3.5PVTTLSDD406 pKa = 3.5VATVTLRR413 pKa = 11.84VTPVAADD420 pKa = 3.67DD421 pKa = 4.98AYY423 pKa = 10.05QTNAEE428 pKa = 4.11STLNVNAPGVFGNDD442 pKa = 3.21TPTSGGTPSVTDD454 pKa = 4.1DD455 pKa = 3.87VDD457 pKa = 5.77DD458 pKa = 4.44GTLTLNDD465 pKa = 4.94DD466 pKa = 3.91GSFTYY471 pKa = 10.43TPDD474 pKa = 3.12AGFAGVDD481 pKa = 3.31TFSYY485 pKa = 10.48SFQSTEE491 pKa = 3.69LDD493 pKa = 3.76SNVATVTITVSPVAVDD509 pKa = 4.49DD510 pKa = 4.25EE511 pKa = 5.26VSAQQCDD518 pKa = 3.62DD519 pKa = 3.57RR520 pKa = 11.84TFTPLANDD528 pKa = 3.84AGVDD532 pKa = 3.72PLGSVPTIIAPPEE545 pKa = 4.61HH546 pKa = 6.37GTLTYY551 pKa = 10.62PEE553 pKa = 4.89DD554 pKa = 3.67LAGGALIYY562 pKa = 11.02DD563 pKa = 4.31PDD565 pKa = 4.96DD566 pKa = 5.11DD567 pKa = 5.27FVGDD571 pKa = 3.82DD572 pKa = 3.29TFTYY576 pKa = 10.07TFATQGVVSNAATVTIHH593 pKa = 5.29VTACEE598 pKa = 4.22SGDD601 pKa = 4.0GGNSDD606 pKa = 5.53DD607 pKa = 5.19GDD609 pKa = 3.7EE610 pKa = 4.66ALPDD614 pKa = 3.73TGSPASPWLLLVAGFATFAGVVLIRR639 pKa = 11.84RR640 pKa = 11.84RR641 pKa = 11.84GGAQAA646 pKa = 4.25
Molecular weight: 66.58 kDa
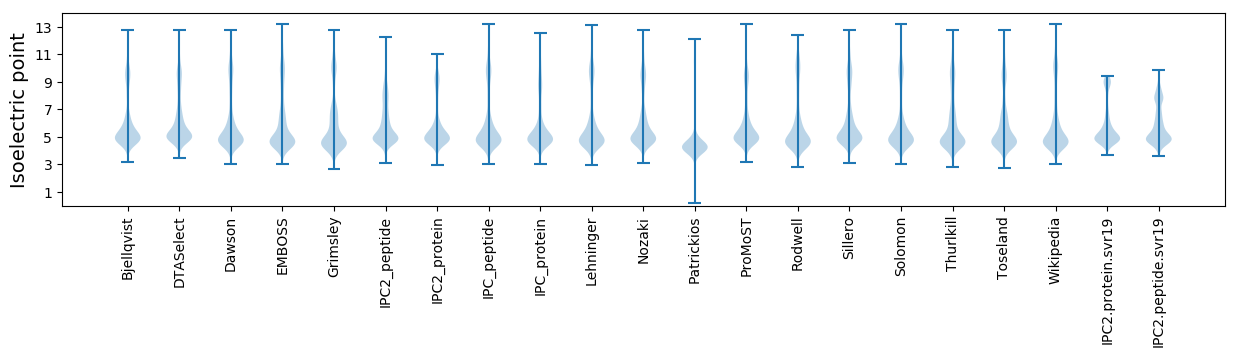
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6V7Y8|A0A0Q6V7Y8_9ACTN Acyl-CoA dehydrogenase OS=Aeromicrobium sp. Root344 OX=1736521 GN=ASC61_07670 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILADD34 pKa = 3.74RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.84GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILADD34 pKa = 3.74RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.84GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1174455 |
37 |
2996 |
314.2 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.547 ± 0.056 | 0.654 ± 0.011 |
6.603 ± 0.033 | 5.602 ± 0.04 |
3.067 ± 0.023 | 8.912 ± 0.033 |
2.168 ± 0.02 | 4.394 ± 0.024 |
2.591 ± 0.035 | 9.98 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.014 | 2.027 ± 0.023 |
5.174 ± 0.031 | 2.815 ± 0.021 |
6.903 ± 0.041 | 5.672 ± 0.029 |
6.281 ± 0.031 | 9.044 ± 0.044 |
1.51 ± 0.018 | 2.087 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
