
Rubrimonas cliftonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rubrimonas
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
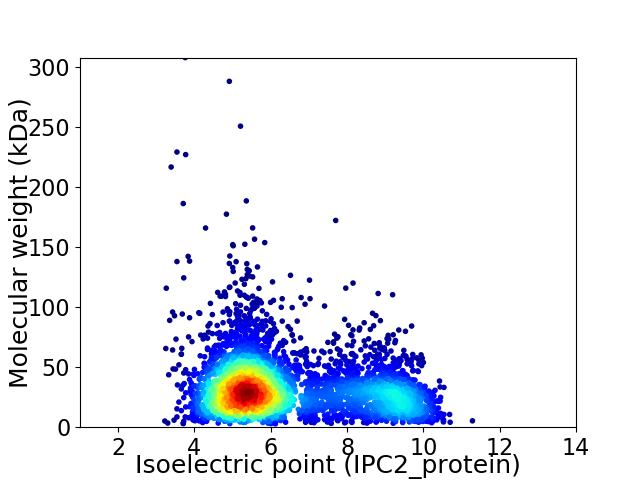
Virtual 2D-PAGE plot for 4574 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3YZ09|A0A1H3YZ09_9RHOB Glycosyltransferase catalytic subunit of cellulose synthase and poly-beta-1 6-N-acetylglucosamine synthase OS=Rubrimonas cliftonensis OX=89524 GN=SAMN05444370_103298 PE=4 SV=1
MM1 pKa = 7.23VSVSHH6 pKa = 6.78AAAAAAVVSMSLAFATSAGAALAPAFVYY34 pKa = 9.59TGSVGVSTDD43 pKa = 3.25GFGSTSGVGTISASAPSGSTVIAAYY68 pKa = 10.15LYY70 pKa = 9.08TSTYY74 pKa = 11.08DD75 pKa = 3.22EE76 pKa = 4.73VGFSTVAPTTVTLDD90 pKa = 3.45GDD92 pKa = 4.03AVTYY96 pKa = 7.4TQSIQNPSYY105 pKa = 11.37SRR107 pKa = 11.84LTAHH111 pKa = 7.23RR112 pKa = 11.84ADD114 pKa = 3.48VTSTVKK120 pKa = 10.43PVIDD124 pKa = 4.51GSAGGVYY131 pKa = 10.44DD132 pKa = 4.56FAYY135 pKa = 10.69NEE137 pKa = 3.96NNAGVEE143 pKa = 4.0NGRR146 pKa = 11.84TGIDD150 pKa = 3.17GSALVVVYY158 pKa = 9.71DD159 pKa = 4.43NPALSDD165 pKa = 3.33RR166 pKa = 11.84TVAVLDD172 pKa = 4.04GFSDD176 pKa = 3.44IAGDD180 pKa = 3.8TFDD183 pKa = 3.52VTFGVPLDD191 pKa = 3.6PTAPGFNLEE200 pKa = 4.07MFLGIGYY207 pKa = 8.26SCCNQASNITVNGTLISRR225 pKa = 11.84NSGNFDD231 pKa = 5.24DD232 pKa = 6.49GLQQANGSLITMGSFDD248 pKa = 4.58DD249 pKa = 4.67APSAFLPSYY258 pKa = 10.64AADD261 pKa = 3.47TEE263 pKa = 4.54RR264 pKa = 11.84YY265 pKa = 10.05DD266 pKa = 3.74LAPYY270 pKa = 10.02VNAGDD275 pKa = 3.95TLLSVDD281 pKa = 4.04TLNPSVNDD289 pKa = 3.83NIFVAVLDD297 pKa = 4.02LTGTASVVNPDD308 pKa = 3.56VPTDD312 pKa = 3.63PTVPPIPLPASMWLLVSGIAMAAGVARR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84ACC343 pKa = 3.8
MM1 pKa = 7.23VSVSHH6 pKa = 6.78AAAAAAVVSMSLAFATSAGAALAPAFVYY34 pKa = 9.59TGSVGVSTDD43 pKa = 3.25GFGSTSGVGTISASAPSGSTVIAAYY68 pKa = 10.15LYY70 pKa = 9.08TSTYY74 pKa = 11.08DD75 pKa = 3.22EE76 pKa = 4.73VGFSTVAPTTVTLDD90 pKa = 3.45GDD92 pKa = 4.03AVTYY96 pKa = 7.4TQSIQNPSYY105 pKa = 11.37SRR107 pKa = 11.84LTAHH111 pKa = 7.23RR112 pKa = 11.84ADD114 pKa = 3.48VTSTVKK120 pKa = 10.43PVIDD124 pKa = 4.51GSAGGVYY131 pKa = 10.44DD132 pKa = 4.56FAYY135 pKa = 10.69NEE137 pKa = 3.96NNAGVEE143 pKa = 4.0NGRR146 pKa = 11.84TGIDD150 pKa = 3.17GSALVVVYY158 pKa = 9.71DD159 pKa = 4.43NPALSDD165 pKa = 3.33RR166 pKa = 11.84TVAVLDD172 pKa = 4.04GFSDD176 pKa = 3.44IAGDD180 pKa = 3.8TFDD183 pKa = 3.52VTFGVPLDD191 pKa = 3.6PTAPGFNLEE200 pKa = 4.07MFLGIGYY207 pKa = 8.26SCCNQASNITVNGTLISRR225 pKa = 11.84NSGNFDD231 pKa = 5.24DD232 pKa = 6.49GLQQANGSLITMGSFDD248 pKa = 4.58DD249 pKa = 4.67APSAFLPSYY258 pKa = 10.64AADD261 pKa = 3.47TEE263 pKa = 4.54RR264 pKa = 11.84YY265 pKa = 10.05DD266 pKa = 3.74LAPYY270 pKa = 10.02VNAGDD275 pKa = 3.95TLLSVDD281 pKa = 4.04TLNPSVNDD289 pKa = 3.83NIFVAVLDD297 pKa = 4.02LTGTASVVNPDD308 pKa = 3.56VPTDD312 pKa = 3.63PTVPPIPLPASMWLLVSGIAMAAGVARR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84ACC343 pKa = 3.8
Molecular weight: 34.93 kDa
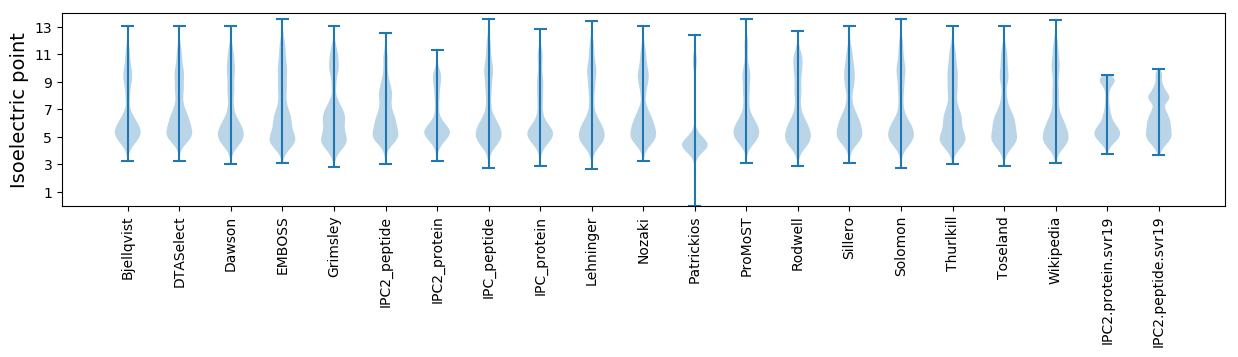
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3ZA24|A0A1H3ZA24_9RHOB Gamma-glutamyl phosphate reductase OS=Rubrimonas cliftonensis OX=89524 GN=proA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.92IVNARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LGRR39 pKa = 11.84KK40 pKa = 8.88HH41 pKa = 5.54LTAA44 pKa = 5.64
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.92IVNARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LGRR39 pKa = 11.84KK40 pKa = 8.88HH41 pKa = 5.54LTAA44 pKa = 5.64
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1452153 |
26 |
3054 |
317.5 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
16.617 ± 0.075 | 0.868 ± 0.012 |
5.827 ± 0.042 | 5.697 ± 0.029 |
3.489 ± 0.025 | 9.413 ± 0.037 |
1.773 ± 0.015 | 3.934 ± 0.025 |
1.951 ± 0.025 | 10.218 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.41 ± 0.017 | 1.855 ± 0.019 |
5.663 ± 0.026 | 2.34 ± 0.017 |
8.122 ± 0.047 | 4.478 ± 0.025 |
4.747 ± 0.025 | 7.41 ± 0.031 |
1.42 ± 0.015 | 1.768 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
