
Beet chlorosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
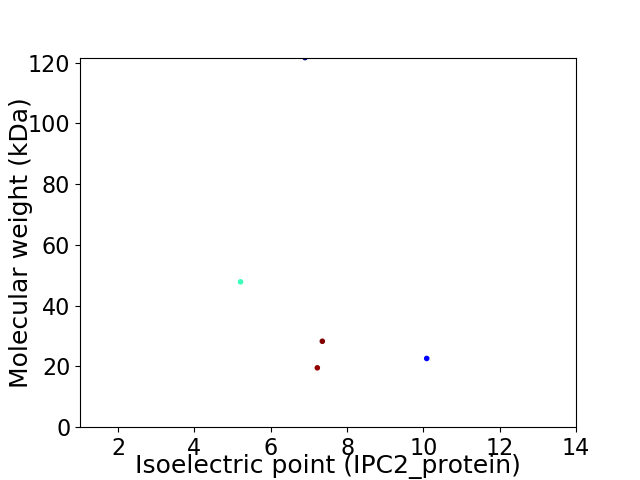
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91AV8|Q91AV8_9LUTE Movement protein OS=Beet chlorosis virus OX=131082 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.23AQSTDD6 pKa = 3.76DD7 pKa = 5.06AISLYY12 pKa = 11.07DD13 pKa = 3.49MPSQRR18 pKa = 11.84FRR20 pKa = 11.84YY21 pKa = 9.81IEE23 pKa = 4.39DD24 pKa = 3.74EE25 pKa = 4.12NMNWTNLDD33 pKa = 3.7SRR35 pKa = 11.84WYY37 pKa = 9.68SQNSLKK43 pKa = 10.38AIPMVIVPVPQGEE56 pKa = 4.3WTVEE60 pKa = 3.43ISMEE64 pKa = 4.75GYY66 pKa = 10.13QPTSSTTDD74 pKa = 3.57PNKK77 pKa = 10.65DD78 pKa = 3.46KK79 pKa = 11.14QDD81 pKa = 3.38GLIAYY86 pKa = 10.04NDD88 pKa = 3.83DD89 pKa = 5.26LKK91 pKa = 11.19EE92 pKa = 3.95GWNVGVYY99 pKa = 10.95NNVEE103 pKa = 3.8ITNNKK108 pKa = 9.19ADD110 pKa = 3.54NTLKK114 pKa = 10.77YY115 pKa = 9.84GHH117 pKa = 7.74PDD119 pKa = 3.01MEE121 pKa = 5.18LNSCHH126 pKa = 7.01FNQQQCLEE134 pKa = 4.15RR135 pKa = 11.84DD136 pKa = 3.51GDD138 pKa = 4.14LTCHH142 pKa = 6.89IKK144 pKa = 8.06TTGDD148 pKa = 3.14NASFFIVGPAVQKK161 pKa = 9.91QSKK164 pKa = 8.52YY165 pKa = 10.45NYY167 pKa = 8.79AVSYY171 pKa = 8.9GAWTDD176 pKa = 3.24RR177 pKa = 11.84MVEE180 pKa = 3.54IGMIAIALDD189 pKa = 3.86EE190 pKa = 4.27QGSSGSARR198 pKa = 11.84TEE200 pKa = 3.59RR201 pKa = 11.84PKK203 pKa = 10.79RR204 pKa = 11.84VGHH207 pKa = 5.9SMAVSTWEE215 pKa = 4.23TINLPEE221 pKa = 6.06KK222 pKa = 10.51EE223 pKa = 4.52DD224 pKa = 3.8SEE226 pKa = 4.47KK227 pKa = 11.19LKK229 pKa = 10.17TGQRR233 pKa = 11.84QDD235 pKa = 3.15LKK237 pKa = 10.52TPFTISGSSDD247 pKa = 2.96VKK249 pKa = 11.02GIEE252 pKa = 4.58KK253 pKa = 10.08RR254 pKa = 11.84DD255 pKa = 3.61LPLPADD261 pKa = 3.75EE262 pKa = 6.24DD263 pKa = 3.75IPDD266 pKa = 5.43FIGNDD271 pKa = 3.0PWSNVSIRR279 pKa = 11.84KK280 pKa = 8.49LQEE283 pKa = 3.6EE284 pKa = 4.31EE285 pKa = 4.52AMTSKK290 pKa = 10.69SGLRR294 pKa = 11.84PQLKK298 pKa = 10.07PPGLPKK304 pKa = 9.66PQPVRR309 pKa = 11.84TIGNFNPTPEE319 pKa = 4.75LVEE322 pKa = 3.88SWRR325 pKa = 11.84PDD327 pKa = 3.39VNPGYY332 pKa = 10.48SKK334 pKa = 11.14EE335 pKa = 4.09DD336 pKa = 3.27VAAATILYY344 pKa = 9.67GGSIKK349 pKa = 10.53DD350 pKa = 3.63GRR352 pKa = 11.84SMIDD356 pKa = 3.06KK357 pKa = 10.07RR358 pKa = 11.84DD359 pKa = 3.47KK360 pKa = 11.16AVLDD364 pKa = 4.22GRR366 pKa = 11.84KK367 pKa = 8.85HH368 pKa = 5.5WGSSLASSLTGGTLKK383 pKa = 10.92ASAKK387 pKa = 9.49SEE389 pKa = 3.99KK390 pKa = 9.94LAKK393 pKa = 9.1LTSRR397 pKa = 11.84EE398 pKa = 3.62RR399 pKa = 11.84AEE401 pKa = 3.8FEE403 pKa = 4.32RR404 pKa = 11.84IKK406 pKa = 10.41RR407 pKa = 11.84QQGTTQASEE416 pKa = 3.97YY417 pKa = 11.04LEE419 pKa = 5.44FILKK423 pKa = 10.35SMNPDD428 pKa = 2.86
MM1 pKa = 7.23AQSTDD6 pKa = 3.76DD7 pKa = 5.06AISLYY12 pKa = 11.07DD13 pKa = 3.49MPSQRR18 pKa = 11.84FRR20 pKa = 11.84YY21 pKa = 9.81IEE23 pKa = 4.39DD24 pKa = 3.74EE25 pKa = 4.12NMNWTNLDD33 pKa = 3.7SRR35 pKa = 11.84WYY37 pKa = 9.68SQNSLKK43 pKa = 10.38AIPMVIVPVPQGEE56 pKa = 4.3WTVEE60 pKa = 3.43ISMEE64 pKa = 4.75GYY66 pKa = 10.13QPTSSTTDD74 pKa = 3.57PNKK77 pKa = 10.65DD78 pKa = 3.46KK79 pKa = 11.14QDD81 pKa = 3.38GLIAYY86 pKa = 10.04NDD88 pKa = 3.83DD89 pKa = 5.26LKK91 pKa = 11.19EE92 pKa = 3.95GWNVGVYY99 pKa = 10.95NNVEE103 pKa = 3.8ITNNKK108 pKa = 9.19ADD110 pKa = 3.54NTLKK114 pKa = 10.77YY115 pKa = 9.84GHH117 pKa = 7.74PDD119 pKa = 3.01MEE121 pKa = 5.18LNSCHH126 pKa = 7.01FNQQQCLEE134 pKa = 4.15RR135 pKa = 11.84DD136 pKa = 3.51GDD138 pKa = 4.14LTCHH142 pKa = 6.89IKK144 pKa = 8.06TTGDD148 pKa = 3.14NASFFIVGPAVQKK161 pKa = 9.91QSKK164 pKa = 8.52YY165 pKa = 10.45NYY167 pKa = 8.79AVSYY171 pKa = 8.9GAWTDD176 pKa = 3.24RR177 pKa = 11.84MVEE180 pKa = 3.54IGMIAIALDD189 pKa = 3.86EE190 pKa = 4.27QGSSGSARR198 pKa = 11.84TEE200 pKa = 3.59RR201 pKa = 11.84PKK203 pKa = 10.79RR204 pKa = 11.84VGHH207 pKa = 5.9SMAVSTWEE215 pKa = 4.23TINLPEE221 pKa = 6.06KK222 pKa = 10.51EE223 pKa = 4.52DD224 pKa = 3.8SEE226 pKa = 4.47KK227 pKa = 11.19LKK229 pKa = 10.17TGQRR233 pKa = 11.84QDD235 pKa = 3.15LKK237 pKa = 10.52TPFTISGSSDD247 pKa = 2.96VKK249 pKa = 11.02GIEE252 pKa = 4.58KK253 pKa = 10.08RR254 pKa = 11.84DD255 pKa = 3.61LPLPADD261 pKa = 3.75EE262 pKa = 6.24DD263 pKa = 3.75IPDD266 pKa = 5.43FIGNDD271 pKa = 3.0PWSNVSIRR279 pKa = 11.84KK280 pKa = 8.49LQEE283 pKa = 3.6EE284 pKa = 4.31EE285 pKa = 4.52AMTSKK290 pKa = 10.69SGLRR294 pKa = 11.84PQLKK298 pKa = 10.07PPGLPKK304 pKa = 9.66PQPVRR309 pKa = 11.84TIGNFNPTPEE319 pKa = 4.75LVEE322 pKa = 3.88SWRR325 pKa = 11.84PDD327 pKa = 3.39VNPGYY332 pKa = 10.48SKK334 pKa = 11.14EE335 pKa = 4.09DD336 pKa = 3.27VAAATILYY344 pKa = 9.67GGSIKK349 pKa = 10.53DD350 pKa = 3.63GRR352 pKa = 11.84SMIDD356 pKa = 3.06KK357 pKa = 10.07RR358 pKa = 11.84DD359 pKa = 3.47KK360 pKa = 11.16AVLDD364 pKa = 4.22GRR366 pKa = 11.84KK367 pKa = 8.85HH368 pKa = 5.5WGSSLASSLTGGTLKK383 pKa = 10.92ASAKK387 pKa = 9.49SEE389 pKa = 3.99KK390 pKa = 9.94LAKK393 pKa = 9.1LTSRR397 pKa = 11.84EE398 pKa = 3.62RR399 pKa = 11.84AEE401 pKa = 3.8FEE403 pKa = 4.32RR404 pKa = 11.84IKK406 pKa = 10.41RR407 pKa = 11.84QQGTTQASEE416 pKa = 3.97YY417 pKa = 11.04LEE419 pKa = 5.44FILKK423 pKa = 10.35SMNPDD428 pKa = 2.86
Molecular weight: 47.83 kDa
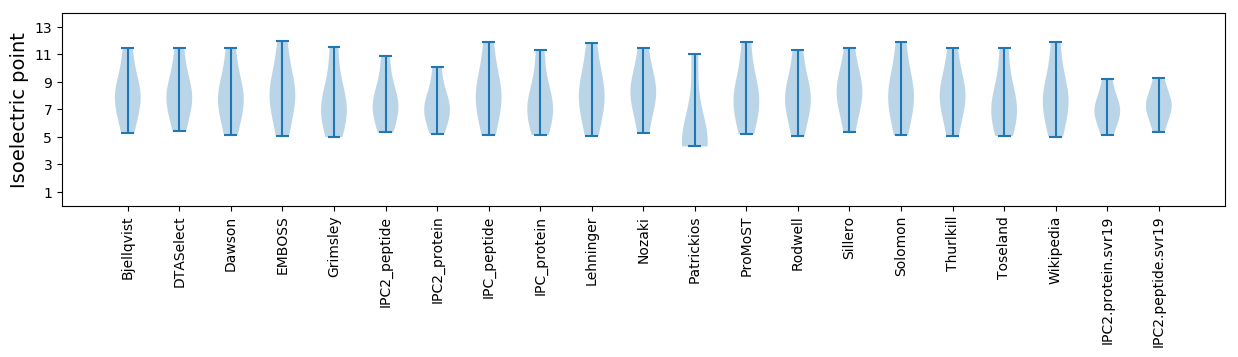
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91AW0|Q91AW0_9LUTE Serine protease OS=Beet chlorosis virus OX=131082 PE=4 SV=1
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QTRR21 pKa = 11.84RR22 pKa = 11.84AQRR25 pKa = 11.84NQPVVVVQTSRR36 pKa = 11.84RR37 pKa = 11.84TQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84AGRR54 pKa = 11.84TVSTRR59 pKa = 11.84GTGQSEE65 pKa = 4.45TFVFSEE71 pKa = 4.69DD72 pKa = 3.37NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGMLKK100 pKa = 10.2AYY102 pKa = 10.09HH103 pKa = 6.71EE104 pKa = 4.71YY105 pKa = 9.92KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.33ASSQSSGSIAYY128 pKa = 9.47EE129 pKa = 3.85LDD131 pKa = 3.29PHH133 pKa = 6.98CKK135 pKa = 10.11LNSLSSTINKK145 pKa = 9.78FGITKK150 pKa = 9.13PGRR153 pKa = 11.84RR154 pKa = 11.84AFTASYY160 pKa = 10.78INGTEE165 pKa = 3.62WHH167 pKa = 7.18DD168 pKa = 3.61VAKK171 pKa = 10.54DD172 pKa = 3.35QFRR175 pKa = 11.84ILYY178 pKa = 8.84KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITIKK195 pKa = 10.75CHH197 pKa = 4.21FHH199 pKa = 6.02NPKK202 pKa = 10.5
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QTRR21 pKa = 11.84RR22 pKa = 11.84AQRR25 pKa = 11.84NQPVVVVQTSRR36 pKa = 11.84RR37 pKa = 11.84TQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84AGRR54 pKa = 11.84TVSTRR59 pKa = 11.84GTGQSEE65 pKa = 4.45TFVFSEE71 pKa = 4.69DD72 pKa = 3.37NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGMLKK100 pKa = 10.2AYY102 pKa = 10.09HH103 pKa = 6.71EE104 pKa = 4.71YY105 pKa = 9.92KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.33ASSQSSGSIAYY128 pKa = 9.47EE129 pKa = 3.85LDD131 pKa = 3.29PHH133 pKa = 6.98CKK135 pKa = 10.11LNSLSSTINKK145 pKa = 9.78FGITKK150 pKa = 9.13PGRR153 pKa = 11.84RR154 pKa = 11.84AFTASYY160 pKa = 10.78INGTEE165 pKa = 3.62WHH167 pKa = 7.18DD168 pKa = 3.61VAKK171 pKa = 10.54DD172 pKa = 3.35QFRR175 pKa = 11.84ILYY178 pKa = 8.84KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITIKK195 pKa = 10.75CHH197 pKa = 4.21FHH199 pKa = 6.02NPKK202 pKa = 10.5
Molecular weight: 22.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2133 |
175 |
1080 |
426.6 |
47.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.391 ± 0.662 | 1.641 ± 0.353 |
5.204 ± 0.766 | 6.142 ± 0.474 |
3.516 ± 0.459 | 6.892 ± 0.521 |
2.532 ± 0.638 | 5.11 ± 0.508 |
5.767 ± 0.682 | 9.048 ± 1.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.28 | 4.594 ± 0.441 |
5.11 ± 0.47 | 3.985 ± 0.326 |
6.423 ± 1.244 | 9.47 ± 0.985 |
6.517 ± 0.591 | 5.579 ± 0.825 |
1.969 ± 0.387 | 2.813 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
