
Devosia sp. I507
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Devosia; unclassified Devosia
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
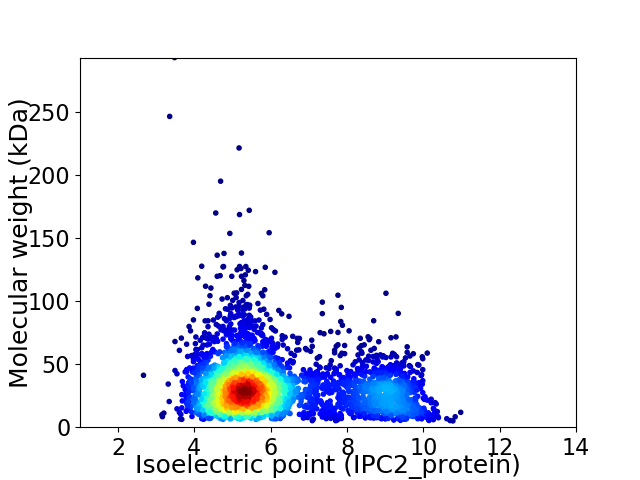
Virtual 2D-PAGE plot for 3776 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L1S497|A0A2L1S497_9RHIZ N-acylglucosamine-6-phosphate 2-epimerase OS=Devosia sp. I507 OX=2083786 GN=C4375_05615 PE=4 SV=1
MM1 pKa = 7.51FFPSDD6 pKa = 3.75PLSEE10 pKa = 4.14IVAHH14 pKa = 5.98FLGYY18 pKa = 9.4FQIAVEE24 pKa = 4.04DD25 pKa = 3.52MKK27 pKa = 11.39VRR29 pKa = 11.84LEE31 pKa = 3.88MALAEE36 pKa = 4.45HH37 pKa = 6.45EE38 pKa = 4.41AEE40 pKa = 4.97AGLEE44 pKa = 4.03APEE47 pKa = 4.03MQQVSVDD54 pKa = 3.71VLQTYY59 pKa = 11.04ALGNYY64 pKa = 9.57APGVFYY70 pKa = 10.74APPEE74 pKa = 3.72FLIRR78 pKa = 11.84GEE80 pKa = 4.37VMFGPTNIGLANTPDD95 pKa = 3.72FTASEE100 pKa = 4.34LDD102 pKa = 3.66SPLSQWPTGPGDD114 pKa = 3.48IVIPLIGTEE123 pKa = 4.15PGSVIAVFSQTISLSDD139 pKa = 3.62DD140 pKa = 3.46DD141 pKa = 4.7VFVMGTLDD149 pKa = 3.66EE150 pKa = 4.54PLVYY154 pKa = 10.44QSGSDD159 pKa = 3.34AGLLGLQAASNAILGPIGSGPGMTSHH185 pKa = 7.0GDD187 pKa = 3.21VPLVMDD193 pKa = 5.24AVHH196 pKa = 6.35ATAKK200 pKa = 10.15AHH202 pKa = 5.89QSNPDD207 pKa = 3.34TQFEE211 pKa = 4.36QVEE214 pKa = 4.86SLTGTYY220 pKa = 10.95VNGKK224 pKa = 9.63LEE226 pKa = 4.22TEE228 pKa = 4.09APEE231 pKa = 4.27LTDD234 pKa = 4.43ALPSHH239 pKa = 6.6LQAEE243 pKa = 4.9AEE245 pKa = 4.35DD246 pKa = 4.68APPPDD251 pKa = 4.19TIKK254 pKa = 9.94VTSEE258 pKa = 4.0HH259 pKa = 6.36EE260 pKa = 3.68AHH262 pKa = 6.4IQIEE266 pKa = 4.1NDD268 pKa = 2.96ATQGSVTYY276 pKa = 10.62NSGGNFLVNEE286 pKa = 4.04AVVLNGGLVGTHH298 pKa = 5.85FVVGGNFHH306 pKa = 6.34QLDD309 pKa = 4.78AIIQTNAFSDD319 pKa = 3.51TDD321 pKa = 3.85YY322 pKa = 11.74VNDD325 pKa = 5.1DD326 pKa = 4.13FPLDD330 pKa = 3.61AQAGGGTKK338 pKa = 10.07AVNVASFVQEE348 pKa = 4.1TFDD351 pKa = 4.08AAGDD355 pKa = 3.74NAEE358 pKa = 4.57ANPGVMPQNWQVTCVSGDD376 pKa = 3.68VIFLEE381 pKa = 4.43WMKK384 pKa = 10.85QLTFMSDD391 pKa = 2.73EE392 pKa = 4.28DD393 pKa = 4.24VGVLTATGSDD403 pKa = 3.12AMVTSGEE410 pKa = 4.23NIGLNSVNFANIGSLFDD427 pKa = 5.1LILVGGNVFDD437 pKa = 5.81ANVIVQNNILYY448 pKa = 10.67DD449 pKa = 3.7NDD451 pKa = 4.5TITMLGAGDD460 pKa = 4.62DD461 pKa = 4.13GDD463 pKa = 4.2YY464 pKa = 10.96SASGSLNTSNNLLWNQASIHH484 pKa = 5.78NVGATEE490 pKa = 4.16VKK492 pKa = 10.52SGVSDD497 pKa = 3.8HH498 pKa = 6.87YY499 pKa = 11.07KK500 pKa = 10.92DD501 pKa = 5.41AMDD504 pKa = 5.06GLANGNKK511 pKa = 9.86SMPASFYY518 pKa = 10.53AAEE521 pKa = 4.46DD522 pKa = 4.02FEE524 pKa = 6.13GIAIPRR530 pKa = 11.84VLFVSGSIYY539 pKa = 10.05DD540 pKa = 3.51LRR542 pKa = 11.84YY543 pKa = 9.69IEE545 pKa = 4.06QTNILGDD552 pKa = 3.72ADD554 pKa = 4.71FVALQKK560 pKa = 10.89KK561 pKa = 9.88AFLDD565 pKa = 4.1GEE567 pKa = 4.58PATEE571 pKa = 4.01WNIDD575 pKa = 3.45TGSNALVNMAAIKK588 pKa = 10.58DD589 pKa = 3.82YY590 pKa = 9.86DD591 pKa = 3.83TFGDD595 pKa = 4.07SIEE598 pKa = 4.37VGGDD602 pKa = 3.18HH603 pKa = 6.77YY604 pKa = 11.49SDD606 pKa = 5.3SILIQANILEE616 pKa = 4.43AQGDD620 pKa = 4.04GDD622 pKa = 5.57DD623 pKa = 5.34DD624 pKa = 4.4DD625 pKa = 6.61ALVSEE630 pKa = 4.99VVAFLDD636 pKa = 3.62HH637 pKa = 6.94TEE639 pKa = 4.2TLADD643 pKa = 3.81TPDD646 pKa = 3.44YY647 pKa = 11.0SALGDD652 pKa = 3.92TTHH655 pKa = 7.69DD656 pKa = 3.77AAPVDD661 pKa = 4.09LMQSVLAA668 pKa = 4.92
MM1 pKa = 7.51FFPSDD6 pKa = 3.75PLSEE10 pKa = 4.14IVAHH14 pKa = 5.98FLGYY18 pKa = 9.4FQIAVEE24 pKa = 4.04DD25 pKa = 3.52MKK27 pKa = 11.39VRR29 pKa = 11.84LEE31 pKa = 3.88MALAEE36 pKa = 4.45HH37 pKa = 6.45EE38 pKa = 4.41AEE40 pKa = 4.97AGLEE44 pKa = 4.03APEE47 pKa = 4.03MQQVSVDD54 pKa = 3.71VLQTYY59 pKa = 11.04ALGNYY64 pKa = 9.57APGVFYY70 pKa = 10.74APPEE74 pKa = 3.72FLIRR78 pKa = 11.84GEE80 pKa = 4.37VMFGPTNIGLANTPDD95 pKa = 3.72FTASEE100 pKa = 4.34LDD102 pKa = 3.66SPLSQWPTGPGDD114 pKa = 3.48IVIPLIGTEE123 pKa = 4.15PGSVIAVFSQTISLSDD139 pKa = 3.62DD140 pKa = 3.46DD141 pKa = 4.7VFVMGTLDD149 pKa = 3.66EE150 pKa = 4.54PLVYY154 pKa = 10.44QSGSDD159 pKa = 3.34AGLLGLQAASNAILGPIGSGPGMTSHH185 pKa = 7.0GDD187 pKa = 3.21VPLVMDD193 pKa = 5.24AVHH196 pKa = 6.35ATAKK200 pKa = 10.15AHH202 pKa = 5.89QSNPDD207 pKa = 3.34TQFEE211 pKa = 4.36QVEE214 pKa = 4.86SLTGTYY220 pKa = 10.95VNGKK224 pKa = 9.63LEE226 pKa = 4.22TEE228 pKa = 4.09APEE231 pKa = 4.27LTDD234 pKa = 4.43ALPSHH239 pKa = 6.6LQAEE243 pKa = 4.9AEE245 pKa = 4.35DD246 pKa = 4.68APPPDD251 pKa = 4.19TIKK254 pKa = 9.94VTSEE258 pKa = 4.0HH259 pKa = 6.36EE260 pKa = 3.68AHH262 pKa = 6.4IQIEE266 pKa = 4.1NDD268 pKa = 2.96ATQGSVTYY276 pKa = 10.62NSGGNFLVNEE286 pKa = 4.04AVVLNGGLVGTHH298 pKa = 5.85FVVGGNFHH306 pKa = 6.34QLDD309 pKa = 4.78AIIQTNAFSDD319 pKa = 3.51TDD321 pKa = 3.85YY322 pKa = 11.74VNDD325 pKa = 5.1DD326 pKa = 4.13FPLDD330 pKa = 3.61AQAGGGTKK338 pKa = 10.07AVNVASFVQEE348 pKa = 4.1TFDD351 pKa = 4.08AAGDD355 pKa = 3.74NAEE358 pKa = 4.57ANPGVMPQNWQVTCVSGDD376 pKa = 3.68VIFLEE381 pKa = 4.43WMKK384 pKa = 10.85QLTFMSDD391 pKa = 2.73EE392 pKa = 4.28DD393 pKa = 4.24VGVLTATGSDD403 pKa = 3.12AMVTSGEE410 pKa = 4.23NIGLNSVNFANIGSLFDD427 pKa = 5.1LILVGGNVFDD437 pKa = 5.81ANVIVQNNILYY448 pKa = 10.67DD449 pKa = 3.7NDD451 pKa = 4.5TITMLGAGDD460 pKa = 4.62DD461 pKa = 4.13GDD463 pKa = 4.2YY464 pKa = 10.96SASGSLNTSNNLLWNQASIHH484 pKa = 5.78NVGATEE490 pKa = 4.16VKK492 pKa = 10.52SGVSDD497 pKa = 3.8HH498 pKa = 6.87YY499 pKa = 11.07KK500 pKa = 10.92DD501 pKa = 5.41AMDD504 pKa = 5.06GLANGNKK511 pKa = 9.86SMPASFYY518 pKa = 10.53AAEE521 pKa = 4.46DD522 pKa = 4.02FEE524 pKa = 6.13GIAIPRR530 pKa = 11.84VLFVSGSIYY539 pKa = 10.05DD540 pKa = 3.51LRR542 pKa = 11.84YY543 pKa = 9.69IEE545 pKa = 4.06QTNILGDD552 pKa = 3.72ADD554 pKa = 4.71FVALQKK560 pKa = 10.89KK561 pKa = 9.88AFLDD565 pKa = 4.1GEE567 pKa = 4.58PATEE571 pKa = 4.01WNIDD575 pKa = 3.45TGSNALVNMAAIKK588 pKa = 10.58DD589 pKa = 3.82YY590 pKa = 9.86DD591 pKa = 3.83TFGDD595 pKa = 4.07SIEE598 pKa = 4.37VGGDD602 pKa = 3.18HH603 pKa = 6.77YY604 pKa = 11.49SDD606 pKa = 5.3SILIQANILEE616 pKa = 4.43AQGDD620 pKa = 4.04GDD622 pKa = 5.57DD623 pKa = 5.34DD624 pKa = 4.4DD625 pKa = 6.61ALVSEE630 pKa = 4.99VVAFLDD636 pKa = 3.62HH637 pKa = 6.94TEE639 pKa = 4.2TLADD643 pKa = 3.81TPDD646 pKa = 3.44YY647 pKa = 11.0SALGDD652 pKa = 3.92TTHH655 pKa = 7.69DD656 pKa = 3.77AAPVDD661 pKa = 4.09LMQSVLAA668 pKa = 4.92
Molecular weight: 70.65 kDa
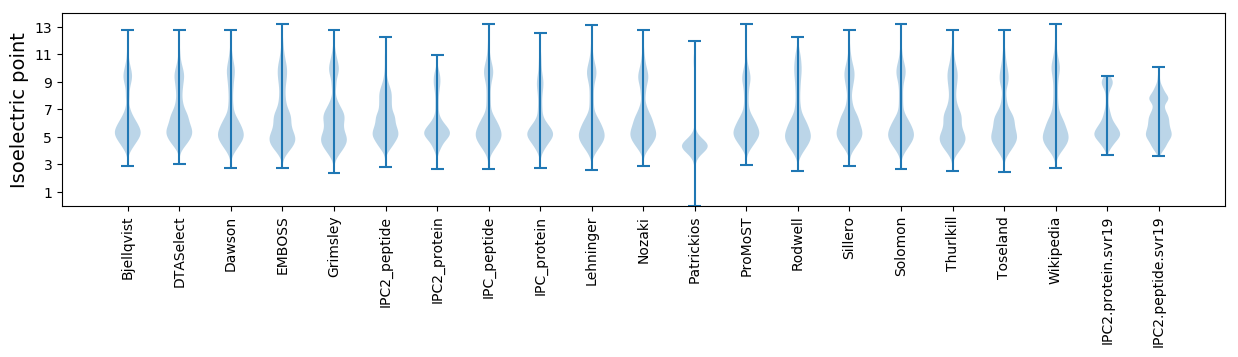
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L1SCC4|A0A2L1SCC4_9RHIZ Choline phosphatase OS=Devosia sp. I507 OX=2083786 GN=cls PE=4 SV=1
MM1 pKa = 6.91TRR3 pKa = 11.84APGRR7 pKa = 11.84PWSRR11 pKa = 11.84PRR13 pKa = 11.84TLRR16 pKa = 11.84PPRR19 pKa = 11.84LRR21 pKa = 11.84TSRR24 pKa = 11.84PARR27 pKa = 11.84PRR29 pKa = 11.84PRR31 pKa = 11.84HH32 pKa = 4.72RR33 pKa = 11.84CQRR36 pKa = 11.84RR37 pKa = 11.84WQRR40 pKa = 11.84PTPRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84LWQSRR51 pKa = 11.84HH52 pKa = 4.56RR53 pKa = 11.84PRR55 pKa = 11.84PKK57 pKa = 9.22RR58 pKa = 11.84WPVGPEE64 pKa = 3.2PGRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84SHH73 pKa = 7.19DD74 pKa = 3.45CLLRR78 pKa = 11.84SPWFRR83 pKa = 11.84LARR86 pKa = 11.84QPPRR90 pKa = 11.84PRR92 pKa = 11.84RR93 pKa = 11.84GG94 pKa = 3.01
MM1 pKa = 6.91TRR3 pKa = 11.84APGRR7 pKa = 11.84PWSRR11 pKa = 11.84PRR13 pKa = 11.84TLRR16 pKa = 11.84PPRR19 pKa = 11.84LRR21 pKa = 11.84TSRR24 pKa = 11.84PARR27 pKa = 11.84PRR29 pKa = 11.84PRR31 pKa = 11.84HH32 pKa = 4.72RR33 pKa = 11.84CQRR36 pKa = 11.84RR37 pKa = 11.84WQRR40 pKa = 11.84PTPRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84LWQSRR51 pKa = 11.84HH52 pKa = 4.56RR53 pKa = 11.84PRR55 pKa = 11.84PKK57 pKa = 9.22RR58 pKa = 11.84WPVGPEE64 pKa = 3.2PGRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84SHH73 pKa = 7.19DD74 pKa = 3.45CLLRR78 pKa = 11.84SPWFRR83 pKa = 11.84LARR86 pKa = 11.84QPPRR90 pKa = 11.84PRR92 pKa = 11.84RR93 pKa = 11.84GG94 pKa = 3.01
Molecular weight: 11.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1170282 |
41 |
2904 |
309.9 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.297 ± 0.058 | 0.692 ± 0.011 |
5.796 ± 0.038 | 5.752 ± 0.037 |
3.834 ± 0.026 | 8.55 ± 0.036 |
2.028 ± 0.019 | 5.488 ± 0.027 |
2.946 ± 0.033 | 10.36 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.606 ± 0.02 | 2.801 ± 0.027 |
5.038 ± 0.03 | 3.34 ± 0.023 |
6.492 ± 0.04 | 5.427 ± 0.029 |
5.46 ± 0.024 | 7.502 ± 0.035 |
1.344 ± 0.013 | 2.247 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
