
Triticum turgidum subsp. durum (Durum wheat) (Triticum durum)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade;
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
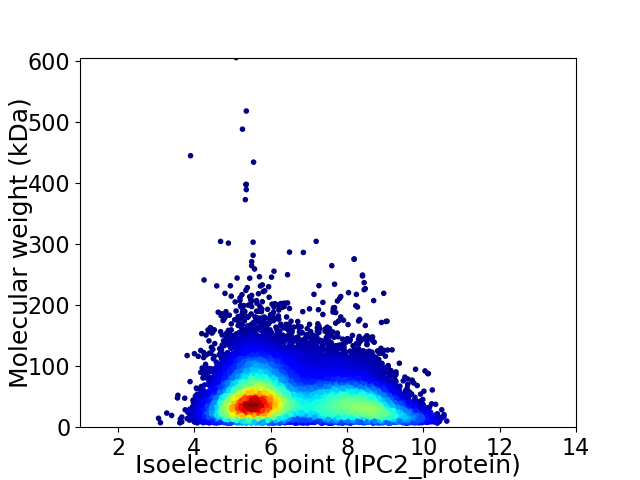
Virtual 2D-PAGE plot for 188121 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A446WTP0|A0A446WTP0_TRITD Isoform of A0A446WTP2 Uncharacterized protein OS=Triticum turgidum subsp. durum OX=4567 GN=TRITD_6Bv1G223910 PE=4 SV=1
MM1 pKa = 7.42SLHH4 pKa = 6.32NSFDD8 pKa = 2.84IMMFILLLGAQDD20 pKa = 3.29MHH22 pKa = 5.84VVCRR26 pKa = 11.84IFQKK30 pKa = 10.73VGSGPQNGAQYY41 pKa = 9.57GAPYY45 pKa = 9.48MEE47 pKa = 5.18EE48 pKa = 3.88EE49 pKa = 4.76WEE51 pKa = 4.61DD52 pKa = 3.53EE53 pKa = 4.22DD54 pKa = 5.85DD55 pKa = 5.58AIEE58 pKa = 4.09NTPTSGTSTEE68 pKa = 4.13MLAITDD74 pKa = 3.86AASAEE79 pKa = 4.5SNVEE83 pKa = 3.92DD84 pKa = 3.37EE85 pKa = 5.45HH86 pKa = 8.41IFSGINEE93 pKa = 4.56FVQTPEE99 pKa = 3.58VLIPQEE105 pKa = 4.07IAPLQAIAPLQVQDD119 pKa = 4.47FNEE122 pKa = 4.32TGMGSYY128 pKa = 11.23ADD130 pKa = 3.79GDD132 pKa = 4.02VPLNEE137 pKa = 4.3ILQEE141 pKa = 4.02PVSDD145 pKa = 3.64VSVDD149 pKa = 3.41NTGEE153 pKa = 4.08PEE155 pKa = 4.15EE156 pKa = 4.08QSPLDD161 pKa = 3.73DD162 pKa = 4.51HH163 pKa = 7.24FSLADD168 pKa = 3.78LSGCHH173 pKa = 5.82NQDD176 pKa = 3.17DD177 pKa = 5.44GYY179 pKa = 11.4VRR181 pKa = 11.84QAGHH185 pKa = 5.92TMWSDD190 pKa = 3.17PSIGDD195 pKa = 3.38QASYY199 pKa = 11.23PMRR202 pKa = 11.84TYY204 pKa = 11.48GNRR207 pKa = 11.84NHH209 pKa = 7.39ANRR212 pKa = 11.84TLSDD216 pKa = 3.57EE217 pKa = 4.33EE218 pKa = 4.9FFDD221 pKa = 4.33TGNDD225 pKa = 3.06ANAYY229 pKa = 9.91SGQQQACPSDD239 pKa = 3.66DD240 pKa = 3.52QNLYY244 pKa = 10.96SGLQQACPSDD254 pKa = 3.84EE255 pKa = 4.17QNLYY259 pKa = 9.9LQPNGLPFPQQLDD272 pKa = 3.79DD273 pKa = 3.79NAPFYY278 pKa = 7.8EE279 pKa = 4.43TSSDD283 pKa = 3.76HH284 pKa = 5.81KK285 pKa = 10.8WEE287 pKa = 4.11VGKK290 pKa = 10.36DD291 pKa = 3.4GYY293 pKa = 11.85VNVDD297 pKa = 4.24DD298 pKa = 5.36ISLFEE303 pKa = 6.42DD304 pKa = 3.98DD305 pKa = 6.57DD306 pKa = 5.12IMALLNASDD315 pKa = 5.71DD316 pKa = 4.15DD317 pKa = 4.49FSSNLLGPVDD327 pKa = 4.33GSNSQLPAASNFDD340 pKa = 3.57QKK342 pKa = 11.64DD343 pKa = 3.39EE344 pKa = 4.54AKK346 pKa = 9.97AQYY349 pKa = 9.62GASSSGSHH357 pKa = 6.17EE358 pKa = 4.16NLYY361 pKa = 9.86PDD363 pKa = 3.68TMVPDD368 pKa = 3.73VPMDD372 pKa = 4.21DD373 pKa = 3.96NAGKK377 pKa = 10.56RR378 pKa = 11.84YY379 pKa = 6.21FTNMLGSYY387 pKa = 7.79PAPPAMASEE396 pKa = 4.84FPPTTGKK403 pKa = 10.59SIAALSGPSQIRR415 pKa = 11.84VTAGIVQLGDD425 pKa = 3.25HH426 pKa = 7.34ADD428 pKa = 4.51NSDD431 pKa = 3.11HH432 pKa = 7.48WPLQKK437 pKa = 10.89NGVFNLLLSFTVEE450 pKa = 4.5SNVSTKK456 pKa = 10.71SITFDD461 pKa = 4.28DD462 pKa = 4.76EE463 pKa = 4.18PATTRR468 pKa = 11.84VSTVPTVLRR477 pKa = 11.84GGLYY481 pKa = 10.61LFFVSAMILMLSFKK495 pKa = 10.49VGSCIYY501 pKa = 10.71SRR503 pKa = 3.98
MM1 pKa = 7.42SLHH4 pKa = 6.32NSFDD8 pKa = 2.84IMMFILLLGAQDD20 pKa = 3.29MHH22 pKa = 5.84VVCRR26 pKa = 11.84IFQKK30 pKa = 10.73VGSGPQNGAQYY41 pKa = 9.57GAPYY45 pKa = 9.48MEE47 pKa = 5.18EE48 pKa = 3.88EE49 pKa = 4.76WEE51 pKa = 4.61DD52 pKa = 3.53EE53 pKa = 4.22DD54 pKa = 5.85DD55 pKa = 5.58AIEE58 pKa = 4.09NTPTSGTSTEE68 pKa = 4.13MLAITDD74 pKa = 3.86AASAEE79 pKa = 4.5SNVEE83 pKa = 3.92DD84 pKa = 3.37EE85 pKa = 5.45HH86 pKa = 8.41IFSGINEE93 pKa = 4.56FVQTPEE99 pKa = 3.58VLIPQEE105 pKa = 4.07IAPLQAIAPLQVQDD119 pKa = 4.47FNEE122 pKa = 4.32TGMGSYY128 pKa = 11.23ADD130 pKa = 3.79GDD132 pKa = 4.02VPLNEE137 pKa = 4.3ILQEE141 pKa = 4.02PVSDD145 pKa = 3.64VSVDD149 pKa = 3.41NTGEE153 pKa = 4.08PEE155 pKa = 4.15EE156 pKa = 4.08QSPLDD161 pKa = 3.73DD162 pKa = 4.51HH163 pKa = 7.24FSLADD168 pKa = 3.78LSGCHH173 pKa = 5.82NQDD176 pKa = 3.17DD177 pKa = 5.44GYY179 pKa = 11.4VRR181 pKa = 11.84QAGHH185 pKa = 5.92TMWSDD190 pKa = 3.17PSIGDD195 pKa = 3.38QASYY199 pKa = 11.23PMRR202 pKa = 11.84TYY204 pKa = 11.48GNRR207 pKa = 11.84NHH209 pKa = 7.39ANRR212 pKa = 11.84TLSDD216 pKa = 3.57EE217 pKa = 4.33EE218 pKa = 4.9FFDD221 pKa = 4.33TGNDD225 pKa = 3.06ANAYY229 pKa = 9.91SGQQQACPSDD239 pKa = 3.66DD240 pKa = 3.52QNLYY244 pKa = 10.96SGLQQACPSDD254 pKa = 3.84EE255 pKa = 4.17QNLYY259 pKa = 9.9LQPNGLPFPQQLDD272 pKa = 3.79DD273 pKa = 3.79NAPFYY278 pKa = 7.8EE279 pKa = 4.43TSSDD283 pKa = 3.76HH284 pKa = 5.81KK285 pKa = 10.8WEE287 pKa = 4.11VGKK290 pKa = 10.36DD291 pKa = 3.4GYY293 pKa = 11.85VNVDD297 pKa = 4.24DD298 pKa = 5.36ISLFEE303 pKa = 6.42DD304 pKa = 3.98DD305 pKa = 6.57DD306 pKa = 5.12IMALLNASDD315 pKa = 5.71DD316 pKa = 4.15DD317 pKa = 4.49FSSNLLGPVDD327 pKa = 4.33GSNSQLPAASNFDD340 pKa = 3.57QKK342 pKa = 11.64DD343 pKa = 3.39EE344 pKa = 4.54AKK346 pKa = 9.97AQYY349 pKa = 9.62GASSSGSHH357 pKa = 6.17EE358 pKa = 4.16NLYY361 pKa = 9.86PDD363 pKa = 3.68TMVPDD368 pKa = 3.73VPMDD372 pKa = 4.21DD373 pKa = 3.96NAGKK377 pKa = 10.56RR378 pKa = 11.84YY379 pKa = 6.21FTNMLGSYY387 pKa = 7.79PAPPAMASEE396 pKa = 4.84FPPTTGKK403 pKa = 10.59SIAALSGPSQIRR415 pKa = 11.84VTAGIVQLGDD425 pKa = 3.25HH426 pKa = 7.34ADD428 pKa = 4.51NSDD431 pKa = 3.11HH432 pKa = 7.48WPLQKK437 pKa = 10.89NGVFNLLLSFTVEE450 pKa = 4.5SNVSTKK456 pKa = 10.71SITFDD461 pKa = 4.28DD462 pKa = 4.76EE463 pKa = 4.18PATTRR468 pKa = 11.84VSTVPTVLRR477 pKa = 11.84GGLYY481 pKa = 10.61LFFVSAMILMLSFKK495 pKa = 10.49VGSCIYY501 pKa = 10.71SRR503 pKa = 3.98
Molecular weight: 54.81 kDa
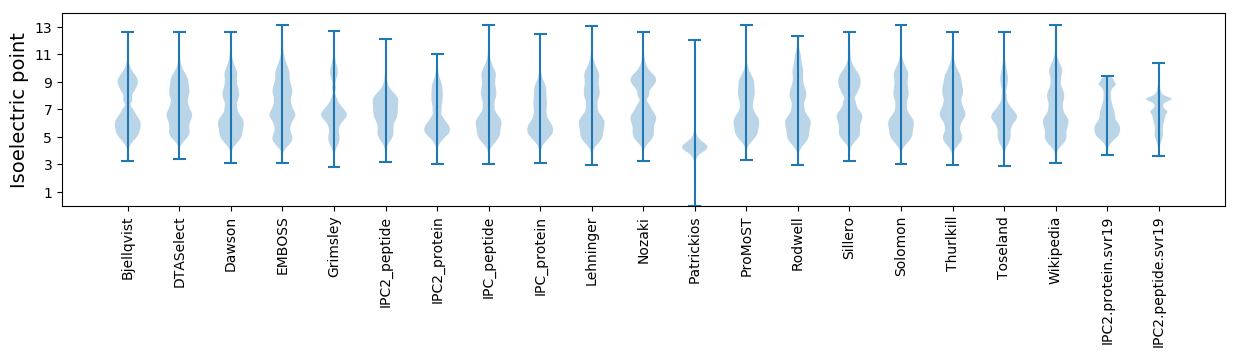
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A446WMR1|A0A446WMR1_TRITD WAT1-related protein OS=Triticum turgidum subsp. durum OX=4567 GN=TRITD_6Bv1G193150 PE=3 SV=1
MM1 pKa = 7.57ARR3 pKa = 11.84TKK5 pKa = 9.25QTARR9 pKa = 11.84KK10 pKa = 7.78STGGKK15 pKa = 9.94APRR18 pKa = 11.84KK19 pKa = 9.29QLATKK24 pKa = 10.05AARR27 pKa = 11.84KK28 pKa = 8.76SAPTTGGVKK37 pKa = 9.88KK38 pKa = 8.99PHH40 pKa = 5.86RR41 pKa = 11.84HH42 pKa = 5.36RR43 pKa = 11.84PGTVALRR50 pKa = 11.84LRR52 pKa = 11.84SASTSRR58 pKa = 11.84ARR60 pKa = 11.84SYY62 pKa = 11.42SSGSFHH68 pKa = 7.51SRR70 pKa = 11.84GLLGRR75 pKa = 11.84LPRR78 pKa = 11.84TSRR81 pKa = 11.84LTSASRR87 pKa = 11.84AMRR90 pKa = 11.84CWPFRR95 pKa = 11.84RR96 pKa = 11.84LQRR99 pKa = 11.84PTWWVSSRR107 pKa = 11.84IRR109 pKa = 11.84GEE111 pKa = 3.85RR112 pKa = 11.84AA113 pKa = 2.7
MM1 pKa = 7.57ARR3 pKa = 11.84TKK5 pKa = 9.25QTARR9 pKa = 11.84KK10 pKa = 7.78STGGKK15 pKa = 9.94APRR18 pKa = 11.84KK19 pKa = 9.29QLATKK24 pKa = 10.05AARR27 pKa = 11.84KK28 pKa = 8.76SAPTTGGVKK37 pKa = 9.88KK38 pKa = 8.99PHH40 pKa = 5.86RR41 pKa = 11.84HH42 pKa = 5.36RR43 pKa = 11.84PGTVALRR50 pKa = 11.84LRR52 pKa = 11.84SASTSRR58 pKa = 11.84ARR60 pKa = 11.84SYY62 pKa = 11.42SSGSFHH68 pKa = 7.51SRR70 pKa = 11.84GLLGRR75 pKa = 11.84LPRR78 pKa = 11.84TSRR81 pKa = 11.84LTSASRR87 pKa = 11.84AMRR90 pKa = 11.84CWPFRR95 pKa = 11.84RR96 pKa = 11.84LQRR99 pKa = 11.84PTWWVSSRR107 pKa = 11.84IRR109 pKa = 11.84GEE111 pKa = 3.85RR112 pKa = 11.84AA113 pKa = 2.7
Molecular weight: 12.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
87439270 |
63 |
5360 |
464.8 |
51.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.309 ± 0.007 | 1.85 ± 0.002 |
5.576 ± 0.004 | 6.227 ± 0.006 |
3.744 ± 0.003 | 7.001 ± 0.006 |
2.471 ± 0.002 | 4.712 ± 0.005 |
5.344 ± 0.005 | 9.671 ± 0.008 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.504 ± 0.002 | 3.775 ± 0.004 |
5.243 ± 0.005 | 3.725 ± 0.004 |
5.786 ± 0.005 | 8.444 ± 0.006 |
4.883 ± 0.003 | 6.747 ± 0.004 |
1.251 ± 0.002 | 2.688 ± 0.003 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
