
Haloplasma contractile SSD-17B
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Haloplasmatales; Haloplasmataceae; Haloplasma; Haloplasma contractile
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
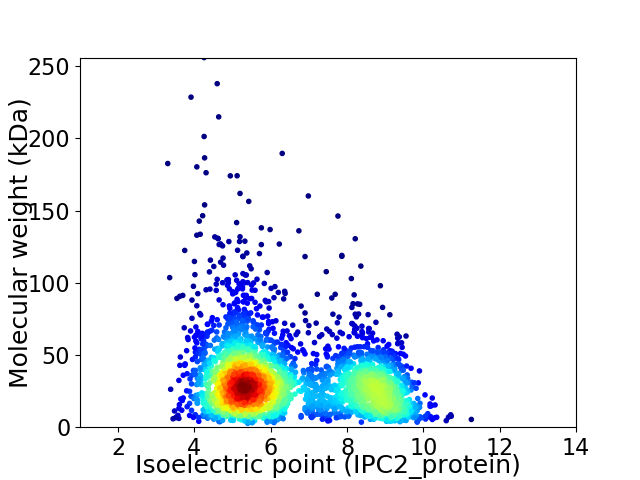
Virtual 2D-PAGE plot for 3019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7PU84|F7PU84_9MOLU Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase OS=Haloplasma contractile SSD-17B OX=1033810 GN=HLPCO_002213 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.24KK3 pKa = 9.38ILSFVFVVTLLAVLGVTSKK22 pKa = 11.64AEE24 pKa = 4.27VIDD27 pKa = 4.41SSTTADD33 pKa = 3.48NVLHH37 pKa = 6.31MKK39 pKa = 10.62DD40 pKa = 3.78DD41 pKa = 4.11AANDD45 pKa = 3.74AVYY48 pKa = 9.9GWSGLKK54 pKa = 9.96IYY56 pKa = 9.96WGTSDD61 pKa = 4.94NDD63 pKa = 3.59PANDD67 pKa = 3.16YY68 pKa = 10.89KK69 pKa = 11.17YY70 pKa = 10.52AYY72 pKa = 9.27KK73 pKa = 10.18VAKK76 pKa = 10.34GNDD79 pKa = 3.61YY80 pKa = 10.98QVDD83 pKa = 3.71QLDD86 pKa = 3.5QDD88 pKa = 3.66GDD90 pKa = 4.03NVVDD94 pKa = 4.55SLPSPKK100 pKa = 9.31WGSFGQVYY108 pKa = 9.97FNFSDD113 pKa = 3.55TDD115 pKa = 3.5IEE117 pKa = 4.26IPRR120 pKa = 11.84ALIGGFMILTVDD132 pKa = 3.33GDD134 pKa = 4.04GNAVRR139 pKa = 11.84FEE141 pKa = 4.38DD142 pKa = 4.48AYY144 pKa = 9.51KK145 pKa = 9.3THH147 pKa = 7.64DD148 pKa = 4.39LGEE151 pKa = 4.7CYY153 pKa = 9.78ATDD156 pKa = 4.04GSIVTYY162 pKa = 9.99TDD164 pKa = 3.0EE165 pKa = 5.24ATDD168 pKa = 3.81CRR170 pKa = 11.84QKK172 pKa = 10.89EE173 pKa = 4.29GNAQAIDD180 pKa = 3.38ASGNLLYY187 pKa = 10.81VDD189 pKa = 4.29ADD191 pKa = 3.91GNEE194 pKa = 4.37TTDD197 pKa = 3.39VTDD200 pKa = 3.41TPKK203 pKa = 10.88AKK205 pKa = 10.15KK206 pKa = 9.77DD207 pKa = 3.66ANDD210 pKa = 3.55EE211 pKa = 4.26NVTGLAWEE219 pKa = 4.4STTDD223 pKa = 3.28GDD225 pKa = 3.93VVYY228 pKa = 10.5IATEE232 pKa = 4.21DD233 pKa = 3.89YY234 pKa = 10.1NTFLTANPTYY244 pKa = 9.95TVVVYY249 pKa = 9.69DD250 pKa = 4.53IIVSNAVAMHH260 pKa = 7.28DD261 pKa = 3.44GTTFTRR267 pKa = 11.84QYY269 pKa = 10.92IDD271 pKa = 3.66FLGNPTDD278 pKa = 3.81NVVVPAGGFVLEE290 pKa = 4.9FEE292 pKa = 4.44WLDD295 pKa = 3.4RR296 pKa = 11.84GHH298 pKa = 6.86FANTNPYY305 pKa = 9.47IQTLFDD311 pKa = 3.47EE312 pKa = 4.78VVNYY316 pKa = 10.91NKK318 pKa = 9.97INYY321 pKa = 9.49SYY323 pKa = 9.91EE324 pKa = 3.81ATVAPTISGVTDD336 pKa = 3.28VNVEE340 pKa = 4.33LGQIKK345 pKa = 10.53SNWLEE350 pKa = 3.98DD351 pKa = 3.81VNASTSQYY359 pKa = 11.04NYY361 pKa = 8.62NTGAMTTIDD370 pKa = 3.54GSVTCTVVEE379 pKa = 4.13SDD381 pKa = 3.42LTEE384 pKa = 4.09VGPCNTTNIDD394 pKa = 3.89TNVMDD399 pKa = 4.44AQYY402 pKa = 11.23KK403 pKa = 9.15LAYY406 pKa = 10.17SVTDD410 pKa = 3.78DD411 pKa = 4.15AGLEE415 pKa = 4.23SVVYY419 pKa = 9.19STVTVVDD426 pKa = 4.02GLAPVISGVDD436 pKa = 3.48NIIVDD441 pKa = 4.15EE442 pKa = 4.63GTDD445 pKa = 3.78LTDD448 pKa = 4.62LLLDD452 pKa = 5.74GITADD457 pKa = 4.53DD458 pKa = 5.26GYY460 pKa = 11.73GNDD463 pKa = 3.01ISEE466 pKa = 5.13NINIVHH472 pKa = 7.61GITDD476 pKa = 3.47VTNPKK481 pKa = 10.32AGTYY485 pKa = 7.48TVTYY489 pKa = 8.95FVYY492 pKa = 9.99NHH494 pKa = 6.39KK495 pKa = 10.53VKK497 pKa = 10.9NDD499 pKa = 3.28FASAVITVNDD509 pKa = 3.48VTNPTVVAGDD519 pKa = 3.2QHH521 pKa = 7.86IDD523 pKa = 3.06QGQEE527 pKa = 3.32FDD529 pKa = 4.09LKK531 pKa = 11.19SGILYY536 pKa = 9.73MSDD539 pKa = 4.22NIDD542 pKa = 3.26NTLDD546 pKa = 3.71VVVTDD551 pKa = 5.5DD552 pKa = 4.79GGFDD556 pKa = 3.72KK557 pKa = 10.55DD558 pKa = 3.57TAGEE562 pKa = 4.13YY563 pKa = 9.32TVAIDD568 pKa = 5.38VYY570 pKa = 10.61DD571 pKa = 3.54QSGNKK576 pKa = 9.98ASTTYY581 pKa = 10.58KK582 pKa = 8.38VTVVAPVEE590 pKa = 4.0IPEE593 pKa = 4.12IPEE596 pKa = 3.66IPEE599 pKa = 3.67IPEE602 pKa = 3.72IPEE605 pKa = 3.87IPEE608 pKa = 4.48DD609 pKa = 3.82SSGLATSGLVVGIVALIVAAGGFVISRR636 pKa = 11.84KK637 pKa = 8.26FF638 pKa = 3.09
MM1 pKa = 7.43KK2 pKa = 10.24KK3 pKa = 9.38ILSFVFVVTLLAVLGVTSKK22 pKa = 11.64AEE24 pKa = 4.27VIDD27 pKa = 4.41SSTTADD33 pKa = 3.48NVLHH37 pKa = 6.31MKK39 pKa = 10.62DD40 pKa = 3.78DD41 pKa = 4.11AANDD45 pKa = 3.74AVYY48 pKa = 9.9GWSGLKK54 pKa = 9.96IYY56 pKa = 9.96WGTSDD61 pKa = 4.94NDD63 pKa = 3.59PANDD67 pKa = 3.16YY68 pKa = 10.89KK69 pKa = 11.17YY70 pKa = 10.52AYY72 pKa = 9.27KK73 pKa = 10.18VAKK76 pKa = 10.34GNDD79 pKa = 3.61YY80 pKa = 10.98QVDD83 pKa = 3.71QLDD86 pKa = 3.5QDD88 pKa = 3.66GDD90 pKa = 4.03NVVDD94 pKa = 4.55SLPSPKK100 pKa = 9.31WGSFGQVYY108 pKa = 9.97FNFSDD113 pKa = 3.55TDD115 pKa = 3.5IEE117 pKa = 4.26IPRR120 pKa = 11.84ALIGGFMILTVDD132 pKa = 3.33GDD134 pKa = 4.04GNAVRR139 pKa = 11.84FEE141 pKa = 4.38DD142 pKa = 4.48AYY144 pKa = 9.51KK145 pKa = 9.3THH147 pKa = 7.64DD148 pKa = 4.39LGEE151 pKa = 4.7CYY153 pKa = 9.78ATDD156 pKa = 4.04GSIVTYY162 pKa = 9.99TDD164 pKa = 3.0EE165 pKa = 5.24ATDD168 pKa = 3.81CRR170 pKa = 11.84QKK172 pKa = 10.89EE173 pKa = 4.29GNAQAIDD180 pKa = 3.38ASGNLLYY187 pKa = 10.81VDD189 pKa = 4.29ADD191 pKa = 3.91GNEE194 pKa = 4.37TTDD197 pKa = 3.39VTDD200 pKa = 3.41TPKK203 pKa = 10.88AKK205 pKa = 10.15KK206 pKa = 9.77DD207 pKa = 3.66ANDD210 pKa = 3.55EE211 pKa = 4.26NVTGLAWEE219 pKa = 4.4STTDD223 pKa = 3.28GDD225 pKa = 3.93VVYY228 pKa = 10.5IATEE232 pKa = 4.21DD233 pKa = 3.89YY234 pKa = 10.1NTFLTANPTYY244 pKa = 9.95TVVVYY249 pKa = 9.69DD250 pKa = 4.53IIVSNAVAMHH260 pKa = 7.28DD261 pKa = 3.44GTTFTRR267 pKa = 11.84QYY269 pKa = 10.92IDD271 pKa = 3.66FLGNPTDD278 pKa = 3.81NVVVPAGGFVLEE290 pKa = 4.9FEE292 pKa = 4.44WLDD295 pKa = 3.4RR296 pKa = 11.84GHH298 pKa = 6.86FANTNPYY305 pKa = 9.47IQTLFDD311 pKa = 3.47EE312 pKa = 4.78VVNYY316 pKa = 10.91NKK318 pKa = 9.97INYY321 pKa = 9.49SYY323 pKa = 9.91EE324 pKa = 3.81ATVAPTISGVTDD336 pKa = 3.28VNVEE340 pKa = 4.33LGQIKK345 pKa = 10.53SNWLEE350 pKa = 3.98DD351 pKa = 3.81VNASTSQYY359 pKa = 11.04NYY361 pKa = 8.62NTGAMTTIDD370 pKa = 3.54GSVTCTVVEE379 pKa = 4.13SDD381 pKa = 3.42LTEE384 pKa = 4.09VGPCNTTNIDD394 pKa = 3.89TNVMDD399 pKa = 4.44AQYY402 pKa = 11.23KK403 pKa = 9.15LAYY406 pKa = 10.17SVTDD410 pKa = 3.78DD411 pKa = 4.15AGLEE415 pKa = 4.23SVVYY419 pKa = 9.19STVTVVDD426 pKa = 4.02GLAPVISGVDD436 pKa = 3.48NIIVDD441 pKa = 4.15EE442 pKa = 4.63GTDD445 pKa = 3.78LTDD448 pKa = 4.62LLLDD452 pKa = 5.74GITADD457 pKa = 4.53DD458 pKa = 5.26GYY460 pKa = 11.73GNDD463 pKa = 3.01ISEE466 pKa = 5.13NINIVHH472 pKa = 7.61GITDD476 pKa = 3.47VTNPKK481 pKa = 10.32AGTYY485 pKa = 7.48TVTYY489 pKa = 8.95FVYY492 pKa = 9.99NHH494 pKa = 6.39KK495 pKa = 10.53VKK497 pKa = 10.9NDD499 pKa = 3.28FASAVITVNDD509 pKa = 3.48VTNPTVVAGDD519 pKa = 3.2QHH521 pKa = 7.86IDD523 pKa = 3.06QGQEE527 pKa = 3.32FDD529 pKa = 4.09LKK531 pKa = 11.19SGILYY536 pKa = 9.73MSDD539 pKa = 4.22NIDD542 pKa = 3.26NTLDD546 pKa = 3.71VVVTDD551 pKa = 5.5DD552 pKa = 4.79GGFDD556 pKa = 3.72KK557 pKa = 10.55DD558 pKa = 3.57TAGEE562 pKa = 4.13YY563 pKa = 9.32TVAIDD568 pKa = 5.38VYY570 pKa = 10.61DD571 pKa = 3.54QSGNKK576 pKa = 9.98ASTTYY581 pKa = 10.58KK582 pKa = 8.38VTVVAPVEE590 pKa = 4.0IPEE593 pKa = 4.12IPEE596 pKa = 3.66IPEE599 pKa = 3.67IPEE602 pKa = 3.72IPEE605 pKa = 3.87IPEE608 pKa = 4.48DD609 pKa = 3.82SSGLATSGLVVGIVALIVAAGGFVISRR636 pKa = 11.84KK637 pKa = 8.26FF638 pKa = 3.09
Molecular weight: 68.84 kDa
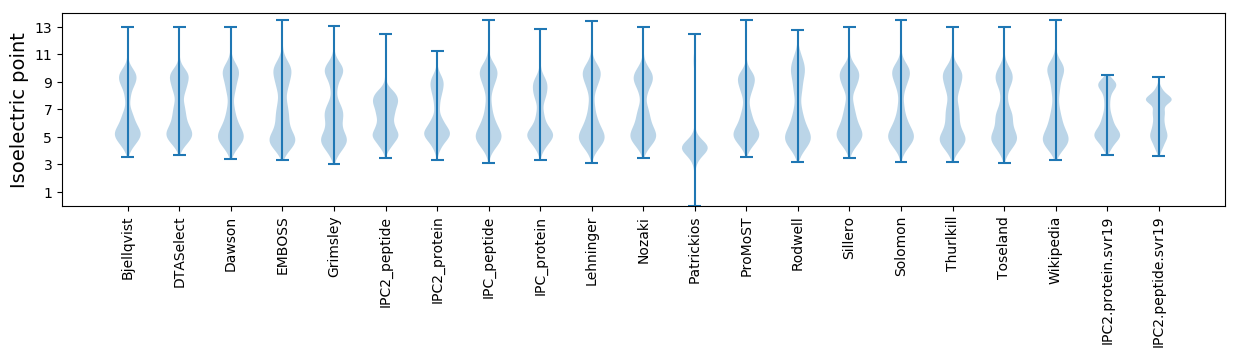
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7PSY9|F7PSY9_9MOLU 50S ribosomal protein L31 OS=Haloplasma contractile SSD-17B OX=1033810 GN=rpmE PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.47RR3 pKa = 11.84TWQPNKK9 pKa = 9.6RR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84NKK14 pKa = 8.11THH16 pKa = 6.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPTGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.41KK37 pKa = 10.29GRR39 pKa = 11.84KK40 pKa = 8.9KK41 pKa = 10.19LTVV44 pKa = 3.1
MM1 pKa = 7.38KK2 pKa = 9.47RR3 pKa = 11.84TWQPNKK9 pKa = 9.6RR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84NKK14 pKa = 8.11THH16 pKa = 6.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPTGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.41KK37 pKa = 10.29GRR39 pKa = 11.84KK40 pKa = 8.9KK41 pKa = 10.19LTVV44 pKa = 3.1
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
927687 |
24 |
2346 |
307.3 |
35.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.062 ± 0.047 | 0.847 ± 0.017 |
6.045 ± 0.044 | 7.14 ± 0.055 |
4.658 ± 0.04 | 5.95 ± 0.048 |
2.041 ± 0.022 | 9.228 ± 0.05 |
7.685 ± 0.056 | 9.626 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.021 | 5.994 ± 0.044 |
2.845 ± 0.027 | 3.03 ± 0.027 |
3.677 ± 0.029 | 5.964 ± 0.033 |
5.726 ± 0.054 | 6.603 ± 0.039 |
0.73 ± 0.015 | 4.714 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
