
Grapevine leafroll-associated virus 3 (isolate United States/NY1) (GLRaV-3) (Grapevine leafroll-associated closterovirus (isolate 109))
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus; Grapevine leafroll-associated virus 3
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
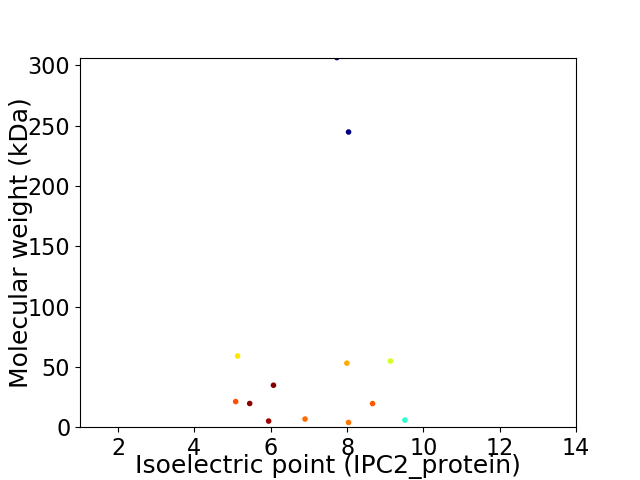
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O71194|P20A_GLRV3 Protein P20A OS=Grapevine leafroll-associated virus 3 (isolate United States/NY1) OX=651354 GN=ORF9 PE=3 SV=1
MM1 pKa = 7.76EE2 pKa = 5.34FRR4 pKa = 11.84PVLITVRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.18PGVNTGSLKK22 pKa = 10.72VIAYY26 pKa = 8.3DD27 pKa = 3.32LHH29 pKa = 7.63YY30 pKa = 11.44DD31 pKa = 4.46NIFDD35 pKa = 3.71NCAVKK40 pKa = 10.48SFRR43 pKa = 11.84DD44 pKa = 3.36TDD46 pKa = 3.53TGFTVMKK53 pKa = 9.95EE54 pKa = 3.97YY55 pKa = 9.76STNSAFILSPYY66 pKa = 10.25KK67 pKa = 10.65LFSAVFNKK75 pKa = 9.74EE76 pKa = 3.29GEE78 pKa = 4.35MISNDD83 pKa = 3.1VGSSFRR89 pKa = 11.84VYY91 pKa = 10.88NIFSQMCKK99 pKa = 10.21DD100 pKa = 3.14INEE103 pKa = 3.82ISEE106 pKa = 4.11IQRR109 pKa = 11.84AGYY112 pKa = 10.35LEE114 pKa = 4.65TYY116 pKa = 10.25LGDD119 pKa = 3.87GQADD123 pKa = 3.38TDD125 pKa = 3.65IFFDD129 pKa = 3.76VLTNNKK135 pKa = 9.96AKK137 pKa = 10.45VRR139 pKa = 11.84WLVNKK144 pKa = 10.06DD145 pKa = 2.9HH146 pKa = 6.72SAWCGILNDD155 pKa = 4.6LKK157 pKa = 10.74WEE159 pKa = 4.06EE160 pKa = 4.25SNKK163 pKa = 10.4EE164 pKa = 3.89KK165 pKa = 10.96FKK167 pKa = 11.24GRR169 pKa = 11.84DD170 pKa = 3.12ILDD173 pKa = 3.85TYY175 pKa = 10.91VLSSDD180 pKa = 3.81YY181 pKa = 11.11PGFKK185 pKa = 10.41
MM1 pKa = 7.76EE2 pKa = 5.34FRR4 pKa = 11.84PVLITVRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.18PGVNTGSLKK22 pKa = 10.72VIAYY26 pKa = 8.3DD27 pKa = 3.32LHH29 pKa = 7.63YY30 pKa = 11.44DD31 pKa = 4.46NIFDD35 pKa = 3.71NCAVKK40 pKa = 10.48SFRR43 pKa = 11.84DD44 pKa = 3.36TDD46 pKa = 3.53TGFTVMKK53 pKa = 9.95EE54 pKa = 3.97YY55 pKa = 9.76STNSAFILSPYY66 pKa = 10.25KK67 pKa = 10.65LFSAVFNKK75 pKa = 9.74EE76 pKa = 3.29GEE78 pKa = 4.35MISNDD83 pKa = 3.1VGSSFRR89 pKa = 11.84VYY91 pKa = 10.88NIFSQMCKK99 pKa = 10.21DD100 pKa = 3.14INEE103 pKa = 3.82ISEE106 pKa = 4.11IQRR109 pKa = 11.84AGYY112 pKa = 10.35LEE114 pKa = 4.65TYY116 pKa = 10.25LGDD119 pKa = 3.87GQADD123 pKa = 3.38TDD125 pKa = 3.65IFFDD129 pKa = 3.76VLTNNKK135 pKa = 9.96AKK137 pKa = 10.45VRR139 pKa = 11.84WLVNKK144 pKa = 10.06DD145 pKa = 2.9HH146 pKa = 6.72SAWCGILNDD155 pKa = 4.6LKK157 pKa = 10.74WEE159 pKa = 4.06EE160 pKa = 4.25SNKK163 pKa = 10.4EE164 pKa = 3.89KK165 pKa = 10.96FKK167 pKa = 11.24GRR169 pKa = 11.84DD170 pKa = 3.12ILDD173 pKa = 3.85TYY175 pKa = 10.91VLSSDD180 pKa = 3.81YY181 pKa = 11.11PGFKK185 pKa = 10.41
Molecular weight: 21.25 kDa
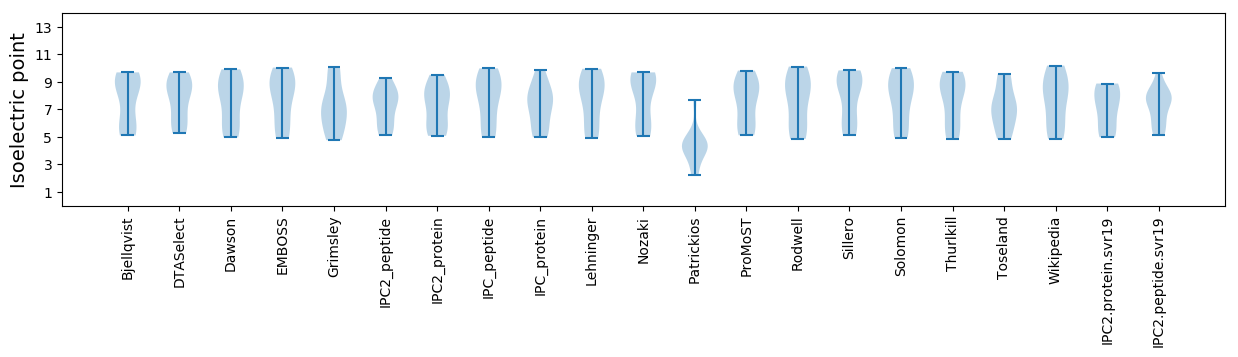
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O41518|CAPSD_GLRV3 Capsid protein OS=Grapevine leafroll-associated virus 3 (isolate United States/NY1) OX=651354 GN=ORF6 PE=3 SV=1
MM1 pKa = 8.07DD2 pKa = 4.6KK3 pKa = 10.7YY4 pKa = 10.96IYY6 pKa = 9.13VTGILNPNEE15 pKa = 3.77ARR17 pKa = 11.84DD18 pKa = 3.75EE19 pKa = 4.32VFSVVNKK26 pKa = 10.52GYY28 pKa = 9.88IGPGGRR34 pKa = 11.84SFSNRR39 pKa = 11.84GSKK42 pKa = 8.45YY43 pKa = 7.87TVVWEE48 pKa = 4.05NSAARR53 pKa = 11.84ISGFTSTSQSTIDD66 pKa = 3.21AFAYY70 pKa = 10.13FLLKK74 pKa = 10.81GGLTTTLSNPINCEE88 pKa = 3.16NWVRR92 pKa = 11.84SSKK95 pKa = 10.71DD96 pKa = 2.89LSAFFRR102 pKa = 11.84TLIKK106 pKa = 10.67GKK108 pKa = 9.7IYY110 pKa = 10.66ASRR113 pKa = 11.84SVDD116 pKa = 3.41SNLPKK121 pKa = 9.96KK122 pKa = 10.69DD123 pKa = 3.69RR124 pKa = 11.84DD125 pKa = 4.38DD126 pKa = 3.37IMEE129 pKa = 4.61ASRR132 pKa = 11.84RR133 pKa = 11.84LSPSDD138 pKa = 3.37AAFCRR143 pKa = 11.84AVSVQVGKK151 pKa = 10.89YY152 pKa = 10.2VDD154 pKa = 3.41VTQNLEE160 pKa = 4.1STIVPLRR167 pKa = 11.84VMEE170 pKa = 4.09IKK172 pKa = 10.2KK173 pKa = 10.19RR174 pKa = 11.84RR175 pKa = 11.84GSAHH179 pKa = 5.4VSLPKK184 pKa = 10.41VVSAYY189 pKa = 10.95VDD191 pKa = 4.5FYY193 pKa = 11.52TNLQEE198 pKa = 4.35LLSDD202 pKa = 3.51EE203 pKa = 4.4VTRR206 pKa = 11.84ARR208 pKa = 11.84TDD210 pKa = 3.3TVSAYY215 pKa = 9.31ATDD218 pKa = 3.48SMAFLVKK225 pKa = 10.01MLPLTARR232 pKa = 11.84EE233 pKa = 3.85QWLKK237 pKa = 11.32DD238 pKa = 3.23VLGYY242 pKa = 10.93LLVRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84PANFSYY254 pKa = 10.49DD255 pKa = 3.43VRR257 pKa = 11.84VAWVYY262 pKa = 11.26DD263 pKa = 3.89VIATLKK269 pKa = 10.67LVIRR273 pKa = 11.84LFFNKK278 pKa = 8.3DD279 pKa = 2.99TPGGIKK285 pKa = 9.87DD286 pKa = 4.75LKK288 pKa = 8.83PCVPIEE294 pKa = 4.24SFDD297 pKa = 3.87PFHH300 pKa = 6.71EE301 pKa = 4.49LSSYY305 pKa = 10.52FSRR308 pKa = 11.84LSYY311 pKa = 11.32EE312 pKa = 3.79MTTGKK317 pKa = 10.25GGKK320 pKa = 8.56ICPEE324 pKa = 3.53IAEE327 pKa = 4.28KK328 pKa = 10.46LVRR331 pKa = 11.84RR332 pKa = 11.84LMEE335 pKa = 4.26EE336 pKa = 3.83NYY338 pKa = 10.34KK339 pKa = 10.78LRR341 pKa = 11.84LTPVMALIIILVYY354 pKa = 9.77YY355 pKa = 10.13SIYY358 pKa = 9.0GTNATRR364 pKa = 11.84IKK366 pKa = 10.37RR367 pKa = 11.84RR368 pKa = 11.84PDD370 pKa = 3.28FLNVRR375 pKa = 11.84IKK377 pKa = 11.08GRR379 pKa = 11.84VEE381 pKa = 3.8KK382 pKa = 10.97VSLRR386 pKa = 11.84GVEE389 pKa = 4.03DD390 pKa = 3.31RR391 pKa = 11.84AFRR394 pKa = 11.84ISEE397 pKa = 3.84KK398 pKa = 10.58RR399 pKa = 11.84GINAQRR405 pKa = 11.84VLCRR409 pKa = 11.84YY410 pKa = 10.02YY411 pKa = 11.36SDD413 pKa = 3.73LTCLARR419 pKa = 11.84RR420 pKa = 11.84HH421 pKa = 5.07YY422 pKa = 10.62GIRR425 pKa = 11.84RR426 pKa = 11.84NNWKK430 pKa = 8.2TLSYY434 pKa = 11.39VDD436 pKa = 3.46GTLAYY441 pKa = 8.18DD442 pKa = 3.61TADD445 pKa = 4.21CITSKK450 pKa = 10.3VRR452 pKa = 11.84NTINTADD459 pKa = 3.63HH460 pKa = 6.8ASIIHH465 pKa = 5.83YY466 pKa = 10.35IKK468 pKa = 10.13TNEE471 pKa = 3.99NQVTGTTLPHH481 pKa = 5.83QLL483 pKa = 3.71
MM1 pKa = 8.07DD2 pKa = 4.6KK3 pKa = 10.7YY4 pKa = 10.96IYY6 pKa = 9.13VTGILNPNEE15 pKa = 3.77ARR17 pKa = 11.84DD18 pKa = 3.75EE19 pKa = 4.32VFSVVNKK26 pKa = 10.52GYY28 pKa = 9.88IGPGGRR34 pKa = 11.84SFSNRR39 pKa = 11.84GSKK42 pKa = 8.45YY43 pKa = 7.87TVVWEE48 pKa = 4.05NSAARR53 pKa = 11.84ISGFTSTSQSTIDD66 pKa = 3.21AFAYY70 pKa = 10.13FLLKK74 pKa = 10.81GGLTTTLSNPINCEE88 pKa = 3.16NWVRR92 pKa = 11.84SSKK95 pKa = 10.71DD96 pKa = 2.89LSAFFRR102 pKa = 11.84TLIKK106 pKa = 10.67GKK108 pKa = 9.7IYY110 pKa = 10.66ASRR113 pKa = 11.84SVDD116 pKa = 3.41SNLPKK121 pKa = 9.96KK122 pKa = 10.69DD123 pKa = 3.69RR124 pKa = 11.84DD125 pKa = 4.38DD126 pKa = 3.37IMEE129 pKa = 4.61ASRR132 pKa = 11.84RR133 pKa = 11.84LSPSDD138 pKa = 3.37AAFCRR143 pKa = 11.84AVSVQVGKK151 pKa = 10.89YY152 pKa = 10.2VDD154 pKa = 3.41VTQNLEE160 pKa = 4.1STIVPLRR167 pKa = 11.84VMEE170 pKa = 4.09IKK172 pKa = 10.2KK173 pKa = 10.19RR174 pKa = 11.84RR175 pKa = 11.84GSAHH179 pKa = 5.4VSLPKK184 pKa = 10.41VVSAYY189 pKa = 10.95VDD191 pKa = 4.5FYY193 pKa = 11.52TNLQEE198 pKa = 4.35LLSDD202 pKa = 3.51EE203 pKa = 4.4VTRR206 pKa = 11.84ARR208 pKa = 11.84TDD210 pKa = 3.3TVSAYY215 pKa = 9.31ATDD218 pKa = 3.48SMAFLVKK225 pKa = 10.01MLPLTARR232 pKa = 11.84EE233 pKa = 3.85QWLKK237 pKa = 11.32DD238 pKa = 3.23VLGYY242 pKa = 10.93LLVRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84PANFSYY254 pKa = 10.49DD255 pKa = 3.43VRR257 pKa = 11.84VAWVYY262 pKa = 11.26DD263 pKa = 3.89VIATLKK269 pKa = 10.67LVIRR273 pKa = 11.84LFFNKK278 pKa = 8.3DD279 pKa = 2.99TPGGIKK285 pKa = 9.87DD286 pKa = 4.75LKK288 pKa = 8.83PCVPIEE294 pKa = 4.24SFDD297 pKa = 3.87PFHH300 pKa = 6.71EE301 pKa = 4.49LSSYY305 pKa = 10.52FSRR308 pKa = 11.84LSYY311 pKa = 11.32EE312 pKa = 3.79MTTGKK317 pKa = 10.25GGKK320 pKa = 8.56ICPEE324 pKa = 3.53IAEE327 pKa = 4.28KK328 pKa = 10.46LVRR331 pKa = 11.84RR332 pKa = 11.84LMEE335 pKa = 4.26EE336 pKa = 3.83NYY338 pKa = 10.34KK339 pKa = 10.78LRR341 pKa = 11.84LTPVMALIIILVYY354 pKa = 9.77YY355 pKa = 10.13SIYY358 pKa = 9.0GTNATRR364 pKa = 11.84IKK366 pKa = 10.37RR367 pKa = 11.84RR368 pKa = 11.84PDD370 pKa = 3.28FLNVRR375 pKa = 11.84IKK377 pKa = 11.08GRR379 pKa = 11.84VEE381 pKa = 3.8KK382 pKa = 10.97VSLRR386 pKa = 11.84GVEE389 pKa = 4.03DD390 pKa = 3.31RR391 pKa = 11.84AFRR394 pKa = 11.84ISEE397 pKa = 3.84KK398 pKa = 10.58RR399 pKa = 11.84GINAQRR405 pKa = 11.84VLCRR409 pKa = 11.84YY410 pKa = 10.02YY411 pKa = 11.36SDD413 pKa = 3.73LTCLARR419 pKa = 11.84RR420 pKa = 11.84HH421 pKa = 5.07YY422 pKa = 10.62GIRR425 pKa = 11.84RR426 pKa = 11.84NNWKK430 pKa = 8.2TLSYY434 pKa = 11.39VDD436 pKa = 3.46GTLAYY441 pKa = 8.18DD442 pKa = 3.61TADD445 pKa = 4.21CITSKK450 pKa = 10.3VRR452 pKa = 11.84NTINTADD459 pKa = 3.63HH460 pKa = 6.8ASIIHH465 pKa = 5.83YY466 pKa = 10.35IKK468 pKa = 10.13TNEE471 pKa = 3.99NQVTGTTLPHH481 pKa = 5.83QLL483 pKa = 3.71
Molecular weight: 54.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7560 |
36 |
2772 |
581.5 |
64.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.222 ± 0.319 | 1.865 ± 0.149 |
5.582 ± 0.247 | 5.146 ± 0.201 |
5.053 ± 0.336 | 7.222 ± 0.438 |
1.574 ± 0.246 | 4.669 ± 0.565 |
5.952 ± 0.253 | 8.452 ± 0.636 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.878 ± 0.157 | 4.021 ± 0.337 |
3.81 ± 0.3 | 2.5 ± 0.24 |
5.952 ± 0.378 | 8.598 ± 0.648 |
6.111 ± 0.315 | 10.04 ± 0.428 |
0.728 ± 0.099 | 3.624 ± 0.309 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
