
Methanobacterium congolense
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanobacterium
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
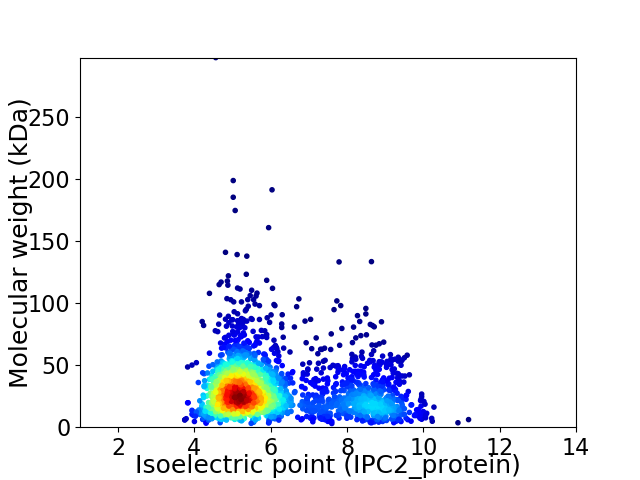
Virtual 2D-PAGE plot for 2340 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
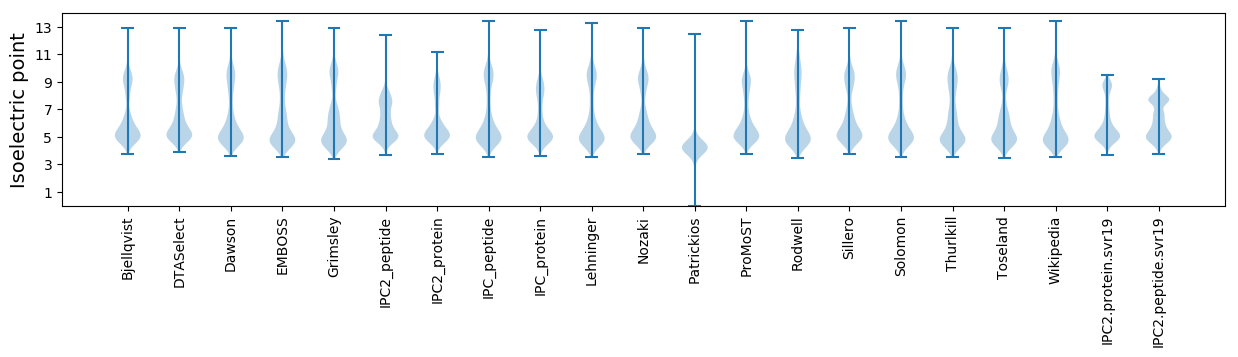
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D3L4I2|A0A1D3L4I2_9EURY Uncharacterized protein OS=Methanobacterium congolense OX=118062 GN=MCBB_1831 PE=4 SV=1
MM1 pKa = 6.73YY2 pKa = 9.63RR3 pKa = 11.84KK4 pKa = 9.35HH5 pKa = 6.49RR6 pKa = 11.84RR7 pKa = 11.84PAFLLLTVFFVICAAGAAAAQEE29 pKa = 4.11AVDD32 pKa = 4.0VNITKK37 pKa = 8.07TAPDD41 pKa = 3.58TVVAGDD47 pKa = 3.86NITYY51 pKa = 10.5NITVTNTNPDD61 pKa = 3.24EE62 pKa = 4.54DD63 pKa = 4.15AQAVTLVDD71 pKa = 4.12DD72 pKa = 4.14VTGKK76 pKa = 10.67LVDD79 pKa = 3.68PQYY82 pKa = 11.62SVDD85 pKa = 4.05GVDD88 pKa = 2.89KK89 pKa = 11.14GSWNGTHH96 pKa = 6.33IWDD99 pKa = 3.91VLPAQTSVLVKK110 pKa = 9.93IWGQVNPSQPAGPWLYY126 pKa = 11.32NDD128 pKa = 3.5ATVFSANDD136 pKa = 3.64PNQDD140 pKa = 2.75NNYY143 pKa = 8.51AHH145 pKa = 6.83TWTLVSTAADD155 pKa = 3.7LNITKK160 pKa = 7.98TAPDD164 pKa = 3.5TVVAGNNITYY174 pKa = 10.25DD175 pKa = 2.93IVVANNGPSDD185 pKa = 3.76AQAVTLVDD193 pKa = 4.12DD194 pKa = 4.18VTGKK198 pKa = 10.76LINPQFSIDD207 pKa = 3.75GVNMGSWGGAYY218 pKa = 9.95GGSYY222 pKa = 9.92VWDD225 pKa = 3.4VLAAQTSVLVKK236 pKa = 9.93IWGQVNPSQPAGPWLYY252 pKa = 11.32NDD254 pKa = 3.35ATVFSVTDD262 pKa = 3.92PNQDD266 pKa = 2.62NNYY269 pKa = 8.42AHH271 pKa = 6.71TWTWVYY277 pKa = 10.15TLADD281 pKa = 3.59VYY283 pKa = 9.96VQKK286 pKa = 9.45TAPANVTAGEE296 pKa = 4.33NITYY300 pKa = 10.44NIVVGNNGPSDD311 pKa = 3.83AQNVTVTDD319 pKa = 4.18TLDD322 pKa = 3.16SWLSGATYY330 pKa = 9.88TLDD333 pKa = 3.76GADD336 pKa = 3.37MGSWTGSLSLGTLAPGKK353 pKa = 9.96YY354 pKa = 6.57VTIIITGLVNATAPAGSLIGNTVSAITDD382 pKa = 3.47TSDD385 pKa = 3.35PFMDD389 pKa = 4.05NNTASTVTGVVTATSGGEE407 pKa = 4.19GVLPPISGGEE417 pKa = 3.95AVTEE421 pKa = 4.21EE422 pKa = 4.37PLSGGEE428 pKa = 3.94AVSAEE433 pKa = 4.62TIPMEE438 pKa = 4.5TTGSPVTLLVMALLVLLASLSGIRR462 pKa = 11.84RR463 pKa = 11.84SS464 pKa = 3.4
MM1 pKa = 6.73YY2 pKa = 9.63RR3 pKa = 11.84KK4 pKa = 9.35HH5 pKa = 6.49RR6 pKa = 11.84RR7 pKa = 11.84PAFLLLTVFFVICAAGAAAAQEE29 pKa = 4.11AVDD32 pKa = 4.0VNITKK37 pKa = 8.07TAPDD41 pKa = 3.58TVVAGDD47 pKa = 3.86NITYY51 pKa = 10.5NITVTNTNPDD61 pKa = 3.24EE62 pKa = 4.54DD63 pKa = 4.15AQAVTLVDD71 pKa = 4.12DD72 pKa = 4.14VTGKK76 pKa = 10.67LVDD79 pKa = 3.68PQYY82 pKa = 11.62SVDD85 pKa = 4.05GVDD88 pKa = 2.89KK89 pKa = 11.14GSWNGTHH96 pKa = 6.33IWDD99 pKa = 3.91VLPAQTSVLVKK110 pKa = 9.93IWGQVNPSQPAGPWLYY126 pKa = 11.32NDD128 pKa = 3.5ATVFSANDD136 pKa = 3.64PNQDD140 pKa = 2.75NNYY143 pKa = 8.51AHH145 pKa = 6.83TWTLVSTAADD155 pKa = 3.7LNITKK160 pKa = 7.98TAPDD164 pKa = 3.5TVVAGNNITYY174 pKa = 10.25DD175 pKa = 2.93IVVANNGPSDD185 pKa = 3.76AQAVTLVDD193 pKa = 4.12DD194 pKa = 4.18VTGKK198 pKa = 10.76LINPQFSIDD207 pKa = 3.75GVNMGSWGGAYY218 pKa = 9.95GGSYY222 pKa = 9.92VWDD225 pKa = 3.4VLAAQTSVLVKK236 pKa = 9.93IWGQVNPSQPAGPWLYY252 pKa = 11.32NDD254 pKa = 3.35ATVFSVTDD262 pKa = 3.92PNQDD266 pKa = 2.62NNYY269 pKa = 8.42AHH271 pKa = 6.71TWTWVYY277 pKa = 10.15TLADD281 pKa = 3.59VYY283 pKa = 9.96VQKK286 pKa = 9.45TAPANVTAGEE296 pKa = 4.33NITYY300 pKa = 10.44NIVVGNNGPSDD311 pKa = 3.83AQNVTVTDD319 pKa = 4.18TLDD322 pKa = 3.16SWLSGATYY330 pKa = 9.88TLDD333 pKa = 3.76GADD336 pKa = 3.37MGSWTGSLSLGTLAPGKK353 pKa = 9.96YY354 pKa = 6.57VTIIITGLVNATAPAGSLIGNTVSAITDD382 pKa = 3.47TSDD385 pKa = 3.35PFMDD389 pKa = 4.05NNTASTVTGVVTATSGGEE407 pKa = 4.19GVLPPISGGEE417 pKa = 3.95AVTEE421 pKa = 4.21EE422 pKa = 4.37PLSGGEE428 pKa = 3.94AVSAEE433 pKa = 4.62TIPMEE438 pKa = 4.5TTGSPVTLLVMALLVLLASLSGIRR462 pKa = 11.84RR463 pKa = 11.84SS464 pKa = 3.4
Molecular weight: 48.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D3L3Z8|A0A1D3L3Z8_9EURY Biopolymer transport protein OS=Methanobacterium congolense OX=118062 GN=MCBB_1823 PE=3 SV=1
MM1 pKa = 7.51SRR3 pKa = 11.84NKK5 pKa = 9.92PLAKK9 pKa = 10.08KK10 pKa = 10.28LRR12 pKa = 11.84LAKK15 pKa = 10.27AGKK18 pKa = 7.75QNRR21 pKa = 11.84RR22 pKa = 11.84VPLWVMLKK30 pKa = 9.19TSRR33 pKa = 11.84KK34 pKa = 8.41VRR36 pKa = 11.84AHH38 pKa = 6.02PKK40 pKa = 7.21MRR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 11.06VKK50 pKa = 10.52AA51 pKa = 3.54
MM1 pKa = 7.51SRR3 pKa = 11.84NKK5 pKa = 9.92PLAKK9 pKa = 10.08KK10 pKa = 10.28LRR12 pKa = 11.84LAKK15 pKa = 10.27AGKK18 pKa = 7.75QNRR21 pKa = 11.84RR22 pKa = 11.84VPLWVMLKK30 pKa = 9.19TSRR33 pKa = 11.84KK34 pKa = 8.41VRR36 pKa = 11.84AHH38 pKa = 6.02PKK40 pKa = 7.21MRR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 11.06VKK50 pKa = 10.52AA51 pKa = 3.54
Molecular weight: 6.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
662468 |
29 |
2816 |
283.1 |
31.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.491 ± 0.059 | 1.197 ± 0.029 |
5.653 ± 0.045 | 7.501 ± 0.084 |
4.181 ± 0.047 | 7.234 ± 0.052 |
1.754 ± 0.023 | 8.057 ± 0.058 |
7.422 ± 0.068 | 9.145 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.928 ± 0.025 | 4.756 ± 0.063 |
3.747 ± 0.034 | 2.339 ± 0.023 |
3.949 ± 0.039 | 6.309 ± 0.047 |
5.414 ± 0.076 | 7.557 ± 0.049 |
0.873 ± 0.019 | 3.495 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
