
Microvirga sp. 17 mud 1-3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Microvirga; unclassified Microvirga
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
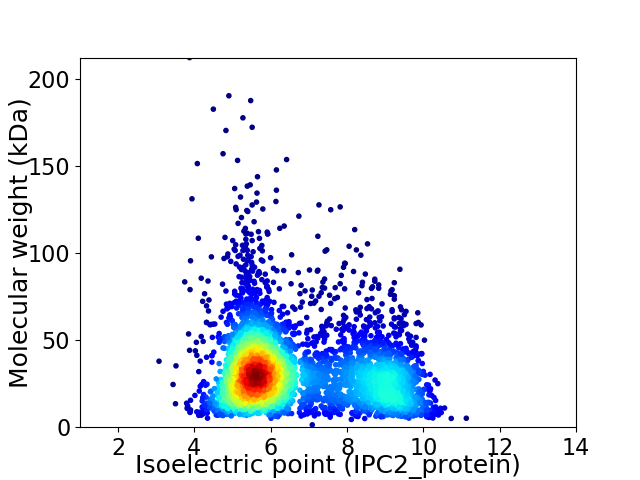
Virtual 2D-PAGE plot for 3992 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z3I9U6|A0A2Z3I9U6_9RHIZ SAM-dependent methyltransferase OS=Microvirga sp. 17 mud 1-3 OX=2082949 GN=C4E04_08670 PE=4 SV=1
MM1 pKa = 8.08SEE3 pKa = 3.87EE4 pKa = 3.99HH5 pKa = 6.88AMATIIGGSGRR16 pKa = 11.84QILKK20 pKa = 8.13GTPSTDD26 pKa = 3.22RR27 pKa = 11.84IFGDD31 pKa = 3.17TSGAYY36 pKa = 9.67SFQSGGDD43 pKa = 3.64DD44 pKa = 4.13KK45 pKa = 11.69LYY47 pKa = 10.79GYY49 pKa = 10.33NGSDD53 pKa = 3.15TLYY56 pKa = 11.34GDD58 pKa = 4.2ALRR61 pKa = 11.84LTGTALGGKK70 pKa = 9.47DD71 pKa = 3.81LFYY74 pKa = 11.15GGNGNDD80 pKa = 4.02TIYY83 pKa = 11.44GDD85 pKa = 3.99ALQLRR90 pKa = 11.84DD91 pKa = 3.81YY92 pKa = 10.25ATGGSDD98 pKa = 3.15TLYY101 pKa = 10.09GQSGNDD107 pKa = 3.62HH108 pKa = 7.13LYY110 pKa = 11.18GDD112 pKa = 4.94AKK114 pKa = 10.9YY115 pKa = 10.36LYY117 pKa = 9.75QKK119 pKa = 10.97AHH121 pKa = 6.1GAGDD125 pKa = 3.74RR126 pKa = 11.84LVGGDD131 pKa = 5.23GIDD134 pKa = 3.29TLYY137 pKa = 11.31GDD139 pKa = 4.61GVALFNYY146 pKa = 10.03AVGGNDD152 pKa = 3.09ILSGDD157 pKa = 3.68RR158 pKa = 11.84GKK160 pKa = 9.61DD161 pKa = 3.12TLYY164 pKa = 10.71GDD166 pKa = 4.31SYY168 pKa = 11.01RR169 pKa = 11.84LEE171 pKa = 4.32GRR173 pKa = 11.84SSGGNDD179 pKa = 3.19SLSGGDD185 pKa = 4.38GVDD188 pKa = 4.23FLYY191 pKa = 11.3GDD193 pKa = 4.31AKK195 pKa = 9.71YY196 pKa = 10.23MKK198 pKa = 9.45YY199 pKa = 10.68ASKK202 pKa = 10.86GGNDD206 pKa = 3.47QLIGGSDD213 pKa = 3.78GDD215 pKa = 4.11SLFGDD220 pKa = 4.54GVRR223 pKa = 11.84MTDD226 pKa = 3.21HH227 pKa = 6.85SVGGSDD233 pKa = 3.51TLFGEE238 pKa = 4.54TGDD241 pKa = 3.82DD242 pKa = 3.47ALYY245 pKa = 11.14GDD247 pKa = 5.06AYY249 pKa = 11.28YY250 pKa = 10.11MGSATHH256 pKa = 6.83GGNDD260 pKa = 3.57SLIGADD266 pKa = 3.91GNDD269 pKa = 3.98TIFGDD274 pKa = 3.98AYY276 pKa = 9.83KK277 pKa = 10.17IEE279 pKa = 4.22YY280 pKa = 9.26NGRR283 pKa = 11.84SGADD287 pKa = 3.43TLSGGAGDD295 pKa = 4.17DD296 pKa = 3.68ALYY299 pKa = 11.12GDD301 pKa = 5.17DD302 pKa = 4.46FQDD305 pKa = 3.3GSTTIISSVLGGNDD319 pKa = 3.94RR320 pKa = 11.84IDD322 pKa = 3.64GGTGNDD328 pKa = 3.76FLAGGLGSDD337 pKa = 3.45VFVFHH342 pKa = 6.97KK343 pKa = 10.15WDD345 pKa = 3.54GRR347 pKa = 11.84DD348 pKa = 3.89VIWDD352 pKa = 3.86FCGTASKK359 pKa = 10.34IPQGDD364 pKa = 4.08KK365 pKa = 10.28IDD367 pKa = 3.92LSDD370 pKa = 2.98WHH372 pKa = 6.39FASFDD377 pKa = 3.33QLTPYY382 pKa = 9.66IHH384 pKa = 6.7YY385 pKa = 10.74NSLDD389 pKa = 3.5TGAVTIDD396 pKa = 3.74LPEE399 pKa = 4.3GQTIILLPVGNNGGIQQLTAADD421 pKa = 5.23FILL424 pKa = 5.1
MM1 pKa = 8.08SEE3 pKa = 3.87EE4 pKa = 3.99HH5 pKa = 6.88AMATIIGGSGRR16 pKa = 11.84QILKK20 pKa = 8.13GTPSTDD26 pKa = 3.22RR27 pKa = 11.84IFGDD31 pKa = 3.17TSGAYY36 pKa = 9.67SFQSGGDD43 pKa = 3.64DD44 pKa = 4.13KK45 pKa = 11.69LYY47 pKa = 10.79GYY49 pKa = 10.33NGSDD53 pKa = 3.15TLYY56 pKa = 11.34GDD58 pKa = 4.2ALRR61 pKa = 11.84LTGTALGGKK70 pKa = 9.47DD71 pKa = 3.81LFYY74 pKa = 11.15GGNGNDD80 pKa = 4.02TIYY83 pKa = 11.44GDD85 pKa = 3.99ALQLRR90 pKa = 11.84DD91 pKa = 3.81YY92 pKa = 10.25ATGGSDD98 pKa = 3.15TLYY101 pKa = 10.09GQSGNDD107 pKa = 3.62HH108 pKa = 7.13LYY110 pKa = 11.18GDD112 pKa = 4.94AKK114 pKa = 10.9YY115 pKa = 10.36LYY117 pKa = 9.75QKK119 pKa = 10.97AHH121 pKa = 6.1GAGDD125 pKa = 3.74RR126 pKa = 11.84LVGGDD131 pKa = 5.23GIDD134 pKa = 3.29TLYY137 pKa = 11.31GDD139 pKa = 4.61GVALFNYY146 pKa = 10.03AVGGNDD152 pKa = 3.09ILSGDD157 pKa = 3.68RR158 pKa = 11.84GKK160 pKa = 9.61DD161 pKa = 3.12TLYY164 pKa = 10.71GDD166 pKa = 4.31SYY168 pKa = 11.01RR169 pKa = 11.84LEE171 pKa = 4.32GRR173 pKa = 11.84SSGGNDD179 pKa = 3.19SLSGGDD185 pKa = 4.38GVDD188 pKa = 4.23FLYY191 pKa = 11.3GDD193 pKa = 4.31AKK195 pKa = 9.71YY196 pKa = 10.23MKK198 pKa = 9.45YY199 pKa = 10.68ASKK202 pKa = 10.86GGNDD206 pKa = 3.47QLIGGSDD213 pKa = 3.78GDD215 pKa = 4.11SLFGDD220 pKa = 4.54GVRR223 pKa = 11.84MTDD226 pKa = 3.21HH227 pKa = 6.85SVGGSDD233 pKa = 3.51TLFGEE238 pKa = 4.54TGDD241 pKa = 3.82DD242 pKa = 3.47ALYY245 pKa = 11.14GDD247 pKa = 5.06AYY249 pKa = 11.28YY250 pKa = 10.11MGSATHH256 pKa = 6.83GGNDD260 pKa = 3.57SLIGADD266 pKa = 3.91GNDD269 pKa = 3.98TIFGDD274 pKa = 3.98AYY276 pKa = 9.83KK277 pKa = 10.17IEE279 pKa = 4.22YY280 pKa = 9.26NGRR283 pKa = 11.84SGADD287 pKa = 3.43TLSGGAGDD295 pKa = 4.17DD296 pKa = 3.68ALYY299 pKa = 11.12GDD301 pKa = 5.17DD302 pKa = 4.46FQDD305 pKa = 3.3GSTTIISSVLGGNDD319 pKa = 3.94RR320 pKa = 11.84IDD322 pKa = 3.64GGTGNDD328 pKa = 3.76FLAGGLGSDD337 pKa = 3.45VFVFHH342 pKa = 6.97KK343 pKa = 10.15WDD345 pKa = 3.54GRR347 pKa = 11.84DD348 pKa = 3.89VIWDD352 pKa = 3.86FCGTASKK359 pKa = 10.34IPQGDD364 pKa = 4.08KK365 pKa = 10.28IDD367 pKa = 3.92LSDD370 pKa = 2.98WHH372 pKa = 6.39FASFDD377 pKa = 3.33QLTPYY382 pKa = 9.66IHH384 pKa = 6.7YY385 pKa = 10.74NSLDD389 pKa = 3.5TGAVTIDD396 pKa = 3.74LPEE399 pKa = 4.3GQTIILLPVGNNGGIQQLTAADD421 pKa = 5.23FILL424 pKa = 5.1
Molecular weight: 44.2 kDa
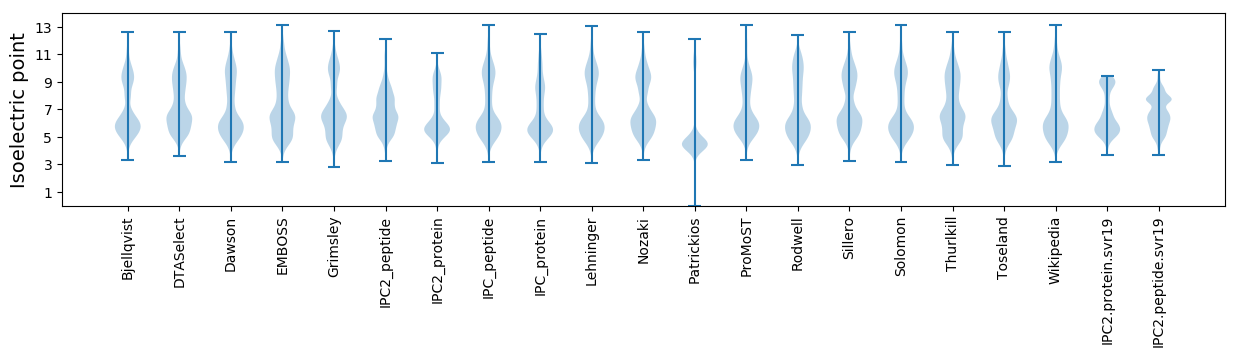
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z3IG99|A0A2Z3IG99_9RHIZ Aminotransferase OS=Microvirga sp. 17 mud 1-3 OX=2082949 GN=C4E04_05440 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.11GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.11GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1263954 |
12 |
2011 |
316.6 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.145 ± 0.047 | 0.77 ± 0.012 |
5.528 ± 0.035 | 5.871 ± 0.035 |
3.715 ± 0.027 | 8.629 ± 0.04 |
1.985 ± 0.02 | 5.171 ± 0.028 |
3.252 ± 0.029 | 10.401 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.363 ± 0.019 | 2.492 ± 0.023 |
5.369 ± 0.034 | 3.074 ± 0.024 |
7.418 ± 0.046 | 5.426 ± 0.025 |
5.315 ± 0.032 | 7.553 ± 0.031 |
1.299 ± 0.014 | 2.225 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
