
Thiogranum longum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Thiogranum
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
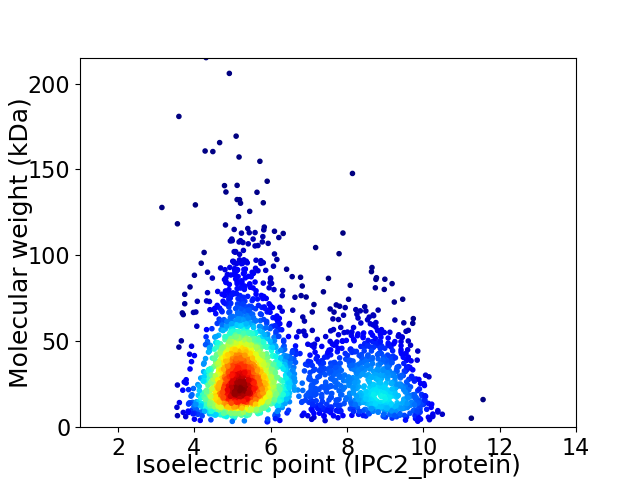
Virtual 2D-PAGE plot for 2795 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1HFB2|A0A4R1HFB2_9GAMM Ankyrin repeat protein OS=Thiogranum longum OX=1537524 GN=DFR30_1308 PE=4 SV=1
MM1 pKa = 7.82HH2 pKa = 7.4SAQCFKK8 pKa = 10.23TKK10 pKa = 10.46KK11 pKa = 9.71RR12 pKa = 11.84ILAAAVLTALSAAAVGPARR31 pKa = 11.84ADD33 pKa = 3.33VQVYY37 pKa = 10.18DD38 pKa = 3.43WTGTFTMINPANVTLVNTDD57 pKa = 2.99VAGNPTKK64 pKa = 10.87GNQTEE69 pKa = 4.34VTGTFTYY76 pKa = 9.38RR77 pKa = 11.84TGSDD81 pKa = 3.32DD82 pKa = 3.74LVNGGGGSGDD92 pKa = 3.27IAIDD96 pKa = 3.3AFNFFGGGPAVAHH109 pKa = 6.99DD110 pKa = 3.92ATFIAIGDD118 pKa = 3.66GHH120 pKa = 6.6GFNGDD125 pKa = 2.87SSGISNGPGDD135 pKa = 4.54LLKK138 pKa = 11.02GDD140 pKa = 5.92LLFDD144 pKa = 3.73WNGTNNIVVEE154 pKa = 4.62IIWNAGGFLTSDD166 pKa = 3.12MGLAGGVGSSVVAGAGAIAPDD187 pKa = 3.75YY188 pKa = 10.68EE189 pKa = 4.61SRR191 pKa = 11.84CVDD194 pKa = 2.91ITFGVCTIALDD205 pKa = 4.12DD206 pKa = 4.27PAAGATEE213 pKa = 5.1WINPADD219 pKa = 3.8GTLVPDD225 pKa = 4.41ASAFGLTTDD234 pKa = 3.61VSGGPILLATVDD246 pKa = 4.24GNPFAQEE253 pKa = 4.18SVDD256 pKa = 3.71PDD258 pKa = 3.94GPGNPIPPVNRR269 pKa = 11.84HH270 pKa = 5.33IDD272 pKa = 3.78GLSGIAMDD280 pKa = 4.54NGPFPGFGANFDD292 pKa = 3.95VRR294 pKa = 11.84TMMLTAFTDD303 pKa = 3.48TTPPVVTLSPNSISINQGDD322 pKa = 4.52AFDD325 pKa = 4.7PLAPPGVTITVVDD338 pKa = 4.3VVDD341 pKa = 3.93GTLDD345 pKa = 3.35VATDD349 pKa = 3.62CSITNGVLTGAAGNYY364 pKa = 6.68TVTYY368 pKa = 9.69NCSDD372 pKa = 3.69LSANANADD380 pKa = 3.58NGSEE384 pKa = 4.1GVLNVQVVAAGAPIITLSGNASVTQEE410 pKa = 4.19CGVAYY415 pKa = 9.2TDD417 pKa = 4.8AGASCTDD424 pKa = 3.34ALLNPITAPADD435 pKa = 3.67FFLDD439 pKa = 4.0TSALNVSTLGVQSVTWSCTDD459 pKa = 2.99TGGGGTTTVDD469 pKa = 2.81RR470 pKa = 11.84VVTVVDD476 pKa = 4.01TSAPVISLNGAASVAIEE493 pKa = 4.25EE494 pKa = 4.64GSVYY498 pKa = 10.51TDD500 pKa = 3.54AGATATDD507 pKa = 3.99GCDD510 pKa = 3.0VGLPALTVVNTVDD523 pKa = 3.18TSVPSTYY530 pKa = 10.68LVTYY534 pKa = 9.86SVSDD538 pKa = 3.45ASGNPATASRR548 pKa = 11.84TVEE551 pKa = 4.13VQPSAPAIVLQGNQTVNLNVGDD573 pKa = 4.07TFDD576 pKa = 4.65PNNPGAVCSDD586 pKa = 3.69AQDD589 pKa = 2.97ATAPVVSVASNVDD602 pKa = 3.18VTQPGTYY609 pKa = 9.17EE610 pKa = 3.93AVYY613 pKa = 10.71SCTDD617 pKa = 2.98TDD619 pKa = 4.03GNTASVTRR627 pKa = 11.84NVIVQGAGFFSAVDD641 pKa = 3.62SNFTMLTPTGDD652 pKa = 3.84FQNGANDD659 pKa = 5.15IVATWDD665 pKa = 3.36GTFNTSVSDD674 pKa = 3.81SNFNMTLATATPTPYY689 pKa = 10.3EE690 pKa = 4.14GSVWTAHH697 pKa = 6.67NIRR700 pKa = 11.84VFGPGSYY707 pKa = 10.05QFDD710 pKa = 3.85VDD712 pKa = 4.07CTPAQIQAGITDD724 pKa = 4.34CPVTDD729 pKa = 3.9LDD731 pKa = 3.97KK732 pKa = 11.52PEE734 pKa = 4.2GRR736 pKa = 11.84VLTLDD741 pKa = 3.36VGPNQLGAHH750 pKa = 6.74ILFDD754 pKa = 3.52WSTTSNIDD762 pKa = 4.59VVNLWNIDD770 pKa = 3.65GAFTSPSGFLGVTGRR785 pKa = 11.84IFNLVSIDD793 pKa = 3.53ANGNNFSGVPMVDD806 pKa = 3.15GPFIGFHH813 pKa = 6.91ANFNLDD819 pKa = 4.0FNPPIQPAAGVRR831 pKa = 11.84APILGAIQNGATRR844 pKa = 11.84RR845 pKa = 11.84AVLVTDD851 pKa = 5.07GIVTVSTDD859 pKa = 2.49IGTTFDD865 pKa = 3.27WSATDD870 pKa = 3.76DD871 pKa = 4.23ALDD874 pKa = 3.99GGATVTTSSLSFDD887 pKa = 4.19PASVAPGFYY896 pKa = 10.23SVGVTIDD903 pKa = 3.5SGLSSEE909 pKa = 4.5ATGSLILEE917 pKa = 4.54VVASVPSVGLDD928 pKa = 3.33DD929 pKa = 4.99SDD931 pKa = 4.51NDD933 pKa = 4.9GIPDD937 pKa = 3.78SVDD940 pKa = 3.14DD941 pKa = 4.37SGPANSLLVDD951 pKa = 4.44AGDD954 pKa = 3.65ATLGSIISSPGTSLSVGNNAYY975 pKa = 9.7IAGIFGGSIPSTDD988 pKa = 3.98LPSDD992 pKa = 3.47SGVEE996 pKa = 4.0EE997 pKa = 4.47SCIGGCFDD1005 pKa = 3.94FQVSGVSPGGSVMVVLPLSEE1025 pKa = 4.48PLPGGSVYY1033 pKa = 10.59RR1034 pKa = 11.84KK1035 pKa = 8.48YY1036 pKa = 10.76NQSAGTWVDD1045 pKa = 3.91FGVGAGNAIASAGKK1059 pKa = 9.73IAGGLCPDD1067 pKa = 4.46EE1068 pKa = 5.68SSASYY1073 pKa = 11.27DD1074 pKa = 3.47DD1075 pKa = 3.67TLGLVAGDD1083 pKa = 3.25EE1084 pKa = 4.8CIRR1087 pKa = 11.84LTIVDD1092 pKa = 4.31GGANDD1097 pKa = 3.81MDD1099 pKa = 4.68SAANPANGIVYY1110 pKa = 10.0DD1111 pKa = 4.15PGGAGVPATPVFASPSTSLGSASGCTVVTAPIDD1144 pKa = 3.42PRR1146 pKa = 11.84QRR1148 pKa = 11.84GDD1150 pKa = 2.44WWLLAMLLGLLGFHH1164 pKa = 6.09AHH1166 pKa = 5.48MRR1168 pKa = 11.84RR1169 pKa = 11.84SRR1171 pKa = 11.84HH1172 pKa = 4.35
MM1 pKa = 7.82HH2 pKa = 7.4SAQCFKK8 pKa = 10.23TKK10 pKa = 10.46KK11 pKa = 9.71RR12 pKa = 11.84ILAAAVLTALSAAAVGPARR31 pKa = 11.84ADD33 pKa = 3.33VQVYY37 pKa = 10.18DD38 pKa = 3.43WTGTFTMINPANVTLVNTDD57 pKa = 2.99VAGNPTKK64 pKa = 10.87GNQTEE69 pKa = 4.34VTGTFTYY76 pKa = 9.38RR77 pKa = 11.84TGSDD81 pKa = 3.32DD82 pKa = 3.74LVNGGGGSGDD92 pKa = 3.27IAIDD96 pKa = 3.3AFNFFGGGPAVAHH109 pKa = 6.99DD110 pKa = 3.92ATFIAIGDD118 pKa = 3.66GHH120 pKa = 6.6GFNGDD125 pKa = 2.87SSGISNGPGDD135 pKa = 4.54LLKK138 pKa = 11.02GDD140 pKa = 5.92LLFDD144 pKa = 3.73WNGTNNIVVEE154 pKa = 4.62IIWNAGGFLTSDD166 pKa = 3.12MGLAGGVGSSVVAGAGAIAPDD187 pKa = 3.75YY188 pKa = 10.68EE189 pKa = 4.61SRR191 pKa = 11.84CVDD194 pKa = 2.91ITFGVCTIALDD205 pKa = 4.12DD206 pKa = 4.27PAAGATEE213 pKa = 5.1WINPADD219 pKa = 3.8GTLVPDD225 pKa = 4.41ASAFGLTTDD234 pKa = 3.61VSGGPILLATVDD246 pKa = 4.24GNPFAQEE253 pKa = 4.18SVDD256 pKa = 3.71PDD258 pKa = 3.94GPGNPIPPVNRR269 pKa = 11.84HH270 pKa = 5.33IDD272 pKa = 3.78GLSGIAMDD280 pKa = 4.54NGPFPGFGANFDD292 pKa = 3.95VRR294 pKa = 11.84TMMLTAFTDD303 pKa = 3.48TTPPVVTLSPNSISINQGDD322 pKa = 4.52AFDD325 pKa = 4.7PLAPPGVTITVVDD338 pKa = 4.3VVDD341 pKa = 3.93GTLDD345 pKa = 3.35VATDD349 pKa = 3.62CSITNGVLTGAAGNYY364 pKa = 6.68TVTYY368 pKa = 9.69NCSDD372 pKa = 3.69LSANANADD380 pKa = 3.58NGSEE384 pKa = 4.1GVLNVQVVAAGAPIITLSGNASVTQEE410 pKa = 4.19CGVAYY415 pKa = 9.2TDD417 pKa = 4.8AGASCTDD424 pKa = 3.34ALLNPITAPADD435 pKa = 3.67FFLDD439 pKa = 4.0TSALNVSTLGVQSVTWSCTDD459 pKa = 2.99TGGGGTTTVDD469 pKa = 2.81RR470 pKa = 11.84VVTVVDD476 pKa = 4.01TSAPVISLNGAASVAIEE493 pKa = 4.25EE494 pKa = 4.64GSVYY498 pKa = 10.51TDD500 pKa = 3.54AGATATDD507 pKa = 3.99GCDD510 pKa = 3.0VGLPALTVVNTVDD523 pKa = 3.18TSVPSTYY530 pKa = 10.68LVTYY534 pKa = 9.86SVSDD538 pKa = 3.45ASGNPATASRR548 pKa = 11.84TVEE551 pKa = 4.13VQPSAPAIVLQGNQTVNLNVGDD573 pKa = 4.07TFDD576 pKa = 4.65PNNPGAVCSDD586 pKa = 3.69AQDD589 pKa = 2.97ATAPVVSVASNVDD602 pKa = 3.18VTQPGTYY609 pKa = 9.17EE610 pKa = 3.93AVYY613 pKa = 10.71SCTDD617 pKa = 2.98TDD619 pKa = 4.03GNTASVTRR627 pKa = 11.84NVIVQGAGFFSAVDD641 pKa = 3.62SNFTMLTPTGDD652 pKa = 3.84FQNGANDD659 pKa = 5.15IVATWDD665 pKa = 3.36GTFNTSVSDD674 pKa = 3.81SNFNMTLATATPTPYY689 pKa = 10.3EE690 pKa = 4.14GSVWTAHH697 pKa = 6.67NIRR700 pKa = 11.84VFGPGSYY707 pKa = 10.05QFDD710 pKa = 3.85VDD712 pKa = 4.07CTPAQIQAGITDD724 pKa = 4.34CPVTDD729 pKa = 3.9LDD731 pKa = 3.97KK732 pKa = 11.52PEE734 pKa = 4.2GRR736 pKa = 11.84VLTLDD741 pKa = 3.36VGPNQLGAHH750 pKa = 6.74ILFDD754 pKa = 3.52WSTTSNIDD762 pKa = 4.59VVNLWNIDD770 pKa = 3.65GAFTSPSGFLGVTGRR785 pKa = 11.84IFNLVSIDD793 pKa = 3.53ANGNNFSGVPMVDD806 pKa = 3.15GPFIGFHH813 pKa = 6.91ANFNLDD819 pKa = 4.0FNPPIQPAAGVRR831 pKa = 11.84APILGAIQNGATRR844 pKa = 11.84RR845 pKa = 11.84AVLVTDD851 pKa = 5.07GIVTVSTDD859 pKa = 2.49IGTTFDD865 pKa = 3.27WSATDD870 pKa = 3.76DD871 pKa = 4.23ALDD874 pKa = 3.99GGATVTTSSLSFDD887 pKa = 4.19PASVAPGFYY896 pKa = 10.23SVGVTIDD903 pKa = 3.5SGLSSEE909 pKa = 4.5ATGSLILEE917 pKa = 4.54VVASVPSVGLDD928 pKa = 3.33DD929 pKa = 4.99SDD931 pKa = 4.51NDD933 pKa = 4.9GIPDD937 pKa = 3.78SVDD940 pKa = 3.14DD941 pKa = 4.37SGPANSLLVDD951 pKa = 4.44AGDD954 pKa = 3.65ATLGSIISSPGTSLSVGNNAYY975 pKa = 9.7IAGIFGGSIPSTDD988 pKa = 3.98LPSDD992 pKa = 3.47SGVEE996 pKa = 4.0EE997 pKa = 4.47SCIGGCFDD1005 pKa = 3.94FQVSGVSPGGSVMVVLPLSEE1025 pKa = 4.48PLPGGSVYY1033 pKa = 10.59RR1034 pKa = 11.84KK1035 pKa = 8.48YY1036 pKa = 10.76NQSAGTWVDD1045 pKa = 3.91FGVGAGNAIASAGKK1059 pKa = 9.73IAGGLCPDD1067 pKa = 4.46EE1068 pKa = 5.68SSASYY1073 pKa = 11.27DD1074 pKa = 3.47DD1075 pKa = 3.67TLGLVAGDD1083 pKa = 3.25EE1084 pKa = 4.8CIRR1087 pKa = 11.84LTIVDD1092 pKa = 4.31GGANDD1097 pKa = 3.81MDD1099 pKa = 4.68SAANPANGIVYY1110 pKa = 10.0DD1111 pKa = 4.15PGGAGVPATPVFASPSTSLGSASGCTVVTAPIDD1144 pKa = 3.42PRR1146 pKa = 11.84QRR1148 pKa = 11.84GDD1150 pKa = 2.44WWLLAMLLGLLGFHH1164 pKa = 6.09AHH1166 pKa = 5.48MRR1168 pKa = 11.84RR1169 pKa = 11.84SRR1171 pKa = 11.84HH1172 pKa = 4.35
Molecular weight: 118.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V2PGH9|A0A4V2PGH9_9GAMM ATP synthase subunit b OS=Thiogranum longum OX=1537524 GN=atpF PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.53NGRR28 pKa = 11.84LVIKK32 pKa = 10.35ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.53NGRR28 pKa = 11.84LVIKK32 pKa = 10.35ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
882141 |
29 |
1966 |
315.6 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.626 ± 0.046 | 1.021 ± 0.018 |
6.0 ± 0.037 | 6.309 ± 0.058 |
3.654 ± 0.031 | 7.763 ± 0.045 |
2.345 ± 0.023 | 5.427 ± 0.037 |
3.837 ± 0.045 | 10.693 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.495 ± 0.025 | 3.29 ± 0.034 |
4.698 ± 0.033 | 3.986 ± 0.034 |
6.603 ± 0.046 | 5.796 ± 0.031 |
5.158 ± 0.043 | 7.325 ± 0.044 |
1.301 ± 0.02 | 2.674 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
