
Beihai sobemo-like virus 27
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
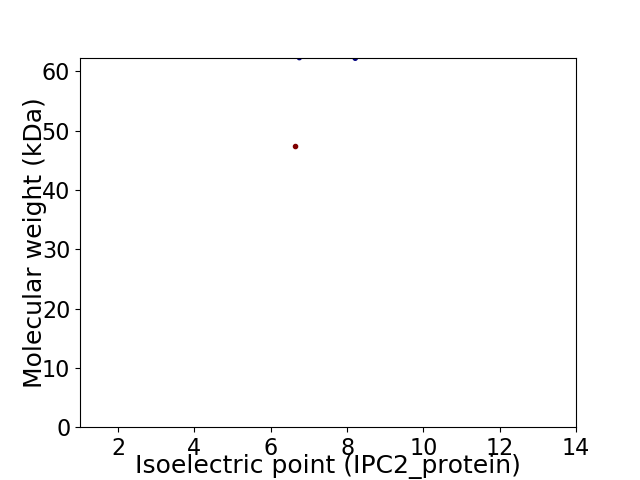
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE72|A0A1L3KE72_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 27 OX=1922699 PE=4 SV=1
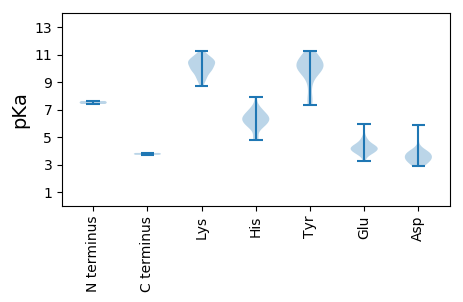
MM1 pKa = 7.62KK2 pKa = 10.44SFAVQSRR9 pKa = 11.84TAHH12 pKa = 5.76IVRR15 pKa = 11.84EE16 pKa = 4.2RR17 pKa = 11.84TISPTDD23 pKa = 3.31EE24 pKa = 4.1EE25 pKa = 4.94LEE27 pKa = 4.7HH28 pKa = 7.91ILYY31 pKa = 9.87HH32 pKa = 6.83AEE34 pKa = 3.83QALAPAKK41 pKa = 9.71WKK43 pKa = 10.77LPSNWNSRR51 pKa = 11.84TAFEE55 pKa = 4.21AALAEE60 pKa = 4.67LDD62 pKa = 3.77WQSSPGYY69 pKa = 8.74PLLRR73 pKa = 11.84EE74 pKa = 4.28APTIGDD80 pKa = 3.37WLLIKK85 pKa = 9.9GTLEE89 pKa = 3.96RR90 pKa = 11.84DD91 pKa = 3.19PSKK94 pKa = 11.26VEE96 pKa = 4.07RR97 pKa = 11.84LWDD100 pKa = 3.57MVQKK104 pKa = 10.28VLKK107 pKa = 10.49GEE109 pKa = 4.49YY110 pKa = 7.59IHH112 pKa = 6.06YY113 pKa = 8.82WRR115 pKa = 11.84VFIKK119 pKa = 10.51SEE121 pKa = 3.49PHH123 pKa = 5.86KK124 pKa = 10.4RR125 pKa = 11.84SKK127 pKa = 10.84ALEE130 pKa = 4.2GRR132 pKa = 11.84WRR134 pKa = 11.84LITAASLPVTVAWMMTFKK152 pKa = 11.02KK153 pKa = 10.83LNDD156 pKa = 3.85RR157 pKa = 11.84LSHH160 pKa = 6.31YY161 pKa = 10.56DD162 pKa = 2.97AALPVQQGYY171 pKa = 7.34VWCSGGWKK179 pKa = 9.58NFKK182 pKa = 10.3RR183 pKa = 11.84RR184 pKa = 11.84IEE186 pKa = 3.97QDD188 pKa = 2.89GLTISVDD195 pKa = 3.04KK196 pKa = 11.04KK197 pKa = 9.77AWDD200 pKa = 3.15WGAPGWAFEE209 pKa = 4.49ADD211 pKa = 3.32LEE213 pKa = 4.58LRR215 pKa = 11.84TRR217 pKa = 11.84LCLNPSPEE225 pKa = 3.73WHH227 pKa = 6.46RR228 pKa = 11.84VATLLYY234 pKa = 10.5ADD236 pKa = 5.05AFQTAKK242 pKa = 10.92LLLPNGIVYY251 pKa = 7.87EE252 pKa = 4.14QQFHH256 pKa = 6.4GFMKK260 pKa = 10.32SGCFNTISTNSNCQILLHH278 pKa = 5.85YY279 pKa = 9.6LAEE282 pKa = 4.22YY283 pKa = 10.04RR284 pKa = 11.84WAKK287 pKa = 10.75AEE289 pKa = 3.91GVSIKK294 pKa = 9.89EE295 pKa = 3.7HH296 pKa = 6.17SKK298 pKa = 10.51ILACGDD304 pKa = 3.37DD305 pKa = 3.86TLQAFYY311 pKa = 10.49TPLYY315 pKa = 10.97GEE317 pKa = 4.9LLEE320 pKa = 4.85QAGCTVKK327 pKa = 10.52EE328 pKa = 4.01AAQNTDD334 pKa = 3.49FMGYY338 pKa = 10.19NYY340 pKa = 10.02DD341 pKa = 3.92GPPEE345 pKa = 3.82PMYY348 pKa = 10.24FAKK351 pKa = 10.65HH352 pKa = 4.96LVKK355 pKa = 10.41FALQKK360 pKa = 9.9PALRR364 pKa = 11.84PEE366 pKa = 4.2TLDD369 pKa = 2.92SYY371 pKa = 10.94MRR373 pKa = 11.84MYY375 pKa = 9.56ATSEE379 pKa = 3.87SRR381 pKa = 11.84HH382 pKa = 4.91FWRR385 pKa = 11.84AVANEE390 pKa = 4.08LEE392 pKa = 4.17IHH394 pKa = 6.35LMSDD398 pKa = 3.21SYY400 pKa = 9.67YY401 pKa = 11.09ARR403 pKa = 11.84WANVPEE409 pKa = 4.24PP410 pKa = 3.84
MM1 pKa = 7.62KK2 pKa = 10.44SFAVQSRR9 pKa = 11.84TAHH12 pKa = 5.76IVRR15 pKa = 11.84EE16 pKa = 4.2RR17 pKa = 11.84TISPTDD23 pKa = 3.31EE24 pKa = 4.1EE25 pKa = 4.94LEE27 pKa = 4.7HH28 pKa = 7.91ILYY31 pKa = 9.87HH32 pKa = 6.83AEE34 pKa = 3.83QALAPAKK41 pKa = 9.71WKK43 pKa = 10.77LPSNWNSRR51 pKa = 11.84TAFEE55 pKa = 4.21AALAEE60 pKa = 4.67LDD62 pKa = 3.77WQSSPGYY69 pKa = 8.74PLLRR73 pKa = 11.84EE74 pKa = 4.28APTIGDD80 pKa = 3.37WLLIKK85 pKa = 9.9GTLEE89 pKa = 3.96RR90 pKa = 11.84DD91 pKa = 3.19PSKK94 pKa = 11.26VEE96 pKa = 4.07RR97 pKa = 11.84LWDD100 pKa = 3.57MVQKK104 pKa = 10.28VLKK107 pKa = 10.49GEE109 pKa = 4.49YY110 pKa = 7.59IHH112 pKa = 6.06YY113 pKa = 8.82WRR115 pKa = 11.84VFIKK119 pKa = 10.51SEE121 pKa = 3.49PHH123 pKa = 5.86KK124 pKa = 10.4RR125 pKa = 11.84SKK127 pKa = 10.84ALEE130 pKa = 4.2GRR132 pKa = 11.84WRR134 pKa = 11.84LITAASLPVTVAWMMTFKK152 pKa = 11.02KK153 pKa = 10.83LNDD156 pKa = 3.85RR157 pKa = 11.84LSHH160 pKa = 6.31YY161 pKa = 10.56DD162 pKa = 2.97AALPVQQGYY171 pKa = 7.34VWCSGGWKK179 pKa = 9.58NFKK182 pKa = 10.3RR183 pKa = 11.84RR184 pKa = 11.84IEE186 pKa = 3.97QDD188 pKa = 2.89GLTISVDD195 pKa = 3.04KK196 pKa = 11.04KK197 pKa = 9.77AWDD200 pKa = 3.15WGAPGWAFEE209 pKa = 4.49ADD211 pKa = 3.32LEE213 pKa = 4.58LRR215 pKa = 11.84TRR217 pKa = 11.84LCLNPSPEE225 pKa = 3.73WHH227 pKa = 6.46RR228 pKa = 11.84VATLLYY234 pKa = 10.5ADD236 pKa = 5.05AFQTAKK242 pKa = 10.92LLLPNGIVYY251 pKa = 7.87EE252 pKa = 4.14QQFHH256 pKa = 6.4GFMKK260 pKa = 10.32SGCFNTISTNSNCQILLHH278 pKa = 5.85YY279 pKa = 9.6LAEE282 pKa = 4.22YY283 pKa = 10.04RR284 pKa = 11.84WAKK287 pKa = 10.75AEE289 pKa = 3.91GVSIKK294 pKa = 9.89EE295 pKa = 3.7HH296 pKa = 6.17SKK298 pKa = 10.51ILACGDD304 pKa = 3.37DD305 pKa = 3.86TLQAFYY311 pKa = 10.49TPLYY315 pKa = 10.97GEE317 pKa = 4.9LLEE320 pKa = 4.85QAGCTVKK327 pKa = 10.52EE328 pKa = 4.01AAQNTDD334 pKa = 3.49FMGYY338 pKa = 10.19NYY340 pKa = 10.02DD341 pKa = 3.92GPPEE345 pKa = 3.82PMYY348 pKa = 10.24FAKK351 pKa = 10.65HH352 pKa = 4.96LVKK355 pKa = 10.41FALQKK360 pKa = 9.9PALRR364 pKa = 11.84PEE366 pKa = 4.2TLDD369 pKa = 2.92SYY371 pKa = 10.94MRR373 pKa = 11.84MYY375 pKa = 9.56ATSEE379 pKa = 3.87SRR381 pKa = 11.84HH382 pKa = 4.91FWRR385 pKa = 11.84AVANEE390 pKa = 4.08LEE392 pKa = 4.17IHH394 pKa = 6.35LMSDD398 pKa = 3.21SYY400 pKa = 9.67YY401 pKa = 11.09ARR403 pKa = 11.84WANVPEE409 pKa = 4.24PP410 pKa = 3.84
Molecular weight: 47.31 kDa
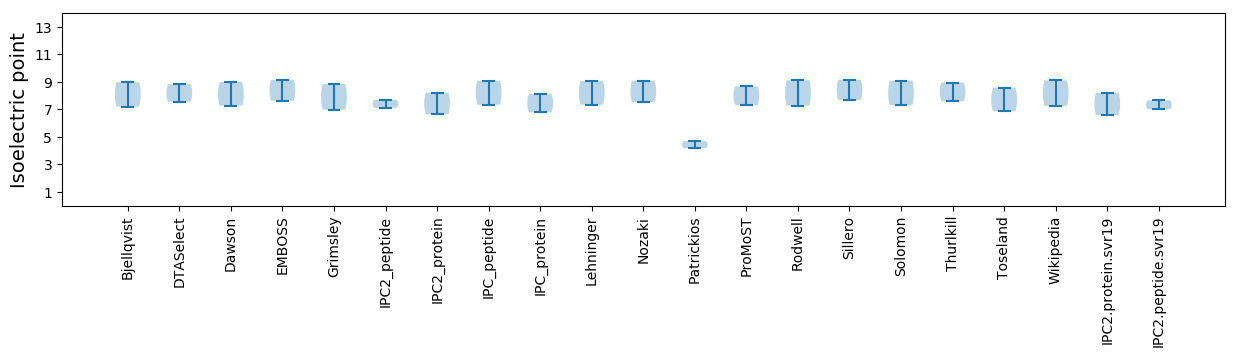
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE72|A0A1L3KE72_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 27 OX=1922699 PE=4 SV=1
MM1 pKa = 7.4EE2 pKa = 4.79FVAKK6 pKa = 10.39ARR8 pKa = 11.84AFLARR13 pKa = 11.84MCLYY17 pKa = 10.22FVGFVVTITNEE28 pKa = 3.72HH29 pKa = 6.07LVALFGAILRR39 pKa = 11.84AVVALFEE46 pKa = 4.29CSMRR50 pKa = 11.84GVHH53 pKa = 6.35HH54 pKa = 6.27MVGALLPEE62 pKa = 4.49ALNPPNRR69 pKa = 11.84KK70 pKa = 9.03NDD72 pKa = 3.65LLRR75 pKa = 11.84QYY77 pKa = 10.13MEE79 pKa = 4.38ANEE82 pKa = 3.99AEE84 pKa = 4.33EE85 pKa = 4.38RR86 pKa = 11.84SRR88 pKa = 11.84SATDD92 pKa = 3.76IIYY95 pKa = 7.62GTSEE99 pKa = 3.69LRR101 pKa = 11.84EE102 pKa = 3.83LHH104 pKa = 6.48MDD106 pKa = 3.97LGYY109 pKa = 10.51QAGFQHH115 pKa = 6.79ASEE118 pKa = 5.12MIISMLALGCLAAWLFSNMPSMNGIGARR146 pKa = 11.84LRR148 pKa = 11.84GCLKK152 pKa = 9.7WRR154 pKa = 11.84PTKK157 pKa = 10.36AVRR160 pKa = 11.84LTDD163 pKa = 3.44VDD165 pKa = 3.74AKK167 pKa = 10.71AAAFATILPEE177 pKa = 3.87TAIPGSNEE185 pKa = 3.27LSMPMTKK192 pKa = 9.94SQVVMFLRR200 pKa = 11.84EE201 pKa = 3.3TDD203 pKa = 3.51AQGKK207 pKa = 8.73NVDD210 pKa = 3.7TVLGNGIRR218 pKa = 11.84FTATRR223 pKa = 11.84LVFPSHH229 pKa = 6.87LGCEE233 pKa = 4.44SFGEE237 pKa = 4.3VWLSSKK243 pKa = 11.0NGGEE247 pKa = 4.08PFCLKK252 pKa = 10.69LGLKK256 pKa = 10.14GDD258 pKa = 3.98YY259 pKa = 10.02QAVAFSNDD267 pKa = 2.88MSFVEE272 pKa = 5.4LPTTVWAGLGVKK284 pKa = 9.55VATIGALSTVGDD296 pKa = 3.75NVTVVGARR304 pKa = 11.84GKK306 pKa = 9.29GTTGVLIAGYY316 pKa = 10.74RR317 pKa = 11.84MGDD320 pKa = 3.41VIYY323 pKa = 11.07NSTTLSGYY331 pKa = 10.33SGAPYY336 pKa = 10.1HH337 pKa = 6.53SGPMTLGVHH346 pKa = 4.82THH348 pKa = 6.44GGKK351 pKa = 10.03QNGGVAATYY360 pKa = 10.1IRR362 pKa = 11.84ARR364 pKa = 11.84LAIYY368 pKa = 9.68DD369 pKa = 4.09ALEE372 pKa = 4.09LDD374 pKa = 3.79IVAEE378 pKa = 4.03SSEE381 pKa = 4.02DD382 pKa = 3.5FMRR385 pKa = 11.84KK386 pKa = 9.11VFDD389 pKa = 3.45SGKK392 pKa = 8.8RR393 pKa = 11.84AHH395 pKa = 6.22RR396 pKa = 11.84TEE398 pKa = 4.33DD399 pKa = 3.13FGDD402 pKa = 3.85FMVVEE407 pKa = 4.6YY408 pKa = 10.39HH409 pKa = 6.59GNHH412 pKa = 5.72HH413 pKa = 5.88TVSKK417 pKa = 10.53QLYY420 pKa = 9.9DD421 pKa = 4.03RR422 pKa = 11.84IKK424 pKa = 9.89SQTYY428 pKa = 9.48HH429 pKa = 7.18PSHH432 pKa = 7.02LSRR435 pKa = 11.84RR436 pKa = 11.84TGLSSSEE443 pKa = 3.68DD444 pKa = 3.3SEE446 pKa = 4.46YY447 pKa = 11.27DD448 pKa = 3.39SEE450 pKa = 5.96SEE452 pKa = 3.95MDD454 pKa = 4.35PPPQLEE460 pKa = 4.22SALNGPVATMAEE472 pKa = 4.3PAVPGYY478 pKa = 10.7DD479 pKa = 3.98DD480 pKa = 5.89GSGLPANIPSSKK492 pKa = 10.09PPSAPPKK499 pKa = 9.44KK500 pKa = 10.01KK501 pKa = 9.97KK502 pKa = 10.04SKK504 pKa = 10.58KK505 pKa = 10.32SGDD508 pKa = 3.57SQRR511 pKa = 11.84ATEE514 pKa = 4.47RR515 pKa = 11.84PTTSRR520 pKa = 11.84KK521 pKa = 9.23SEE523 pKa = 3.95KK524 pKa = 9.28TSSGGSQSRR533 pKa = 11.84KK534 pKa = 9.35SNPSRR539 pKa = 11.84HH540 pKa = 5.77KK541 pKa = 10.48AVKK544 pKa = 9.77RR545 pKa = 11.84DD546 pKa = 2.86IMTKK550 pKa = 10.46FSEE553 pKa = 4.2LTPTHH558 pKa = 7.07RR559 pKa = 11.84ARR561 pKa = 11.84LLTQMAQSLAKK572 pKa = 10.03PP573 pKa = 3.71
MM1 pKa = 7.4EE2 pKa = 4.79FVAKK6 pKa = 10.39ARR8 pKa = 11.84AFLARR13 pKa = 11.84MCLYY17 pKa = 10.22FVGFVVTITNEE28 pKa = 3.72HH29 pKa = 6.07LVALFGAILRR39 pKa = 11.84AVVALFEE46 pKa = 4.29CSMRR50 pKa = 11.84GVHH53 pKa = 6.35HH54 pKa = 6.27MVGALLPEE62 pKa = 4.49ALNPPNRR69 pKa = 11.84KK70 pKa = 9.03NDD72 pKa = 3.65LLRR75 pKa = 11.84QYY77 pKa = 10.13MEE79 pKa = 4.38ANEE82 pKa = 3.99AEE84 pKa = 4.33EE85 pKa = 4.38RR86 pKa = 11.84SRR88 pKa = 11.84SATDD92 pKa = 3.76IIYY95 pKa = 7.62GTSEE99 pKa = 3.69LRR101 pKa = 11.84EE102 pKa = 3.83LHH104 pKa = 6.48MDD106 pKa = 3.97LGYY109 pKa = 10.51QAGFQHH115 pKa = 6.79ASEE118 pKa = 5.12MIISMLALGCLAAWLFSNMPSMNGIGARR146 pKa = 11.84LRR148 pKa = 11.84GCLKK152 pKa = 9.7WRR154 pKa = 11.84PTKK157 pKa = 10.36AVRR160 pKa = 11.84LTDD163 pKa = 3.44VDD165 pKa = 3.74AKK167 pKa = 10.71AAAFATILPEE177 pKa = 3.87TAIPGSNEE185 pKa = 3.27LSMPMTKK192 pKa = 9.94SQVVMFLRR200 pKa = 11.84EE201 pKa = 3.3TDD203 pKa = 3.51AQGKK207 pKa = 8.73NVDD210 pKa = 3.7TVLGNGIRR218 pKa = 11.84FTATRR223 pKa = 11.84LVFPSHH229 pKa = 6.87LGCEE233 pKa = 4.44SFGEE237 pKa = 4.3VWLSSKK243 pKa = 11.0NGGEE247 pKa = 4.08PFCLKK252 pKa = 10.69LGLKK256 pKa = 10.14GDD258 pKa = 3.98YY259 pKa = 10.02QAVAFSNDD267 pKa = 2.88MSFVEE272 pKa = 5.4LPTTVWAGLGVKK284 pKa = 9.55VATIGALSTVGDD296 pKa = 3.75NVTVVGARR304 pKa = 11.84GKK306 pKa = 9.29GTTGVLIAGYY316 pKa = 10.74RR317 pKa = 11.84MGDD320 pKa = 3.41VIYY323 pKa = 11.07NSTTLSGYY331 pKa = 10.33SGAPYY336 pKa = 10.1HH337 pKa = 6.53SGPMTLGVHH346 pKa = 4.82THH348 pKa = 6.44GGKK351 pKa = 10.03QNGGVAATYY360 pKa = 10.1IRR362 pKa = 11.84ARR364 pKa = 11.84LAIYY368 pKa = 9.68DD369 pKa = 4.09ALEE372 pKa = 4.09LDD374 pKa = 3.79IVAEE378 pKa = 4.03SSEE381 pKa = 4.02DD382 pKa = 3.5FMRR385 pKa = 11.84KK386 pKa = 9.11VFDD389 pKa = 3.45SGKK392 pKa = 8.8RR393 pKa = 11.84AHH395 pKa = 6.22RR396 pKa = 11.84TEE398 pKa = 4.33DD399 pKa = 3.13FGDD402 pKa = 3.85FMVVEE407 pKa = 4.6YY408 pKa = 10.39HH409 pKa = 6.59GNHH412 pKa = 5.72HH413 pKa = 5.88TVSKK417 pKa = 10.53QLYY420 pKa = 9.9DD421 pKa = 4.03RR422 pKa = 11.84IKK424 pKa = 9.89SQTYY428 pKa = 9.48HH429 pKa = 7.18PSHH432 pKa = 7.02LSRR435 pKa = 11.84RR436 pKa = 11.84TGLSSSEE443 pKa = 3.68DD444 pKa = 3.3SEE446 pKa = 4.46YY447 pKa = 11.27DD448 pKa = 3.39SEE450 pKa = 5.96SEE452 pKa = 3.95MDD454 pKa = 4.35PPPQLEE460 pKa = 4.22SALNGPVATMAEE472 pKa = 4.3PAVPGYY478 pKa = 10.7DD479 pKa = 3.98DD480 pKa = 5.89GSGLPANIPSSKK492 pKa = 10.09PPSAPPKK499 pKa = 9.44KK500 pKa = 10.01KK501 pKa = 9.97KK502 pKa = 10.04SKK504 pKa = 10.58KK505 pKa = 10.32SGDD508 pKa = 3.57SQRR511 pKa = 11.84ATEE514 pKa = 4.47RR515 pKa = 11.84PTTSRR520 pKa = 11.84KK521 pKa = 9.23SEE523 pKa = 3.95KK524 pKa = 9.28TSSGGSQSRR533 pKa = 11.84KK534 pKa = 9.35SNPSRR539 pKa = 11.84HH540 pKa = 5.77KK541 pKa = 10.48AVKK544 pKa = 9.77RR545 pKa = 11.84DD546 pKa = 2.86IMTKK550 pKa = 10.46FSEE553 pKa = 4.2LTPTHH558 pKa = 7.07RR559 pKa = 11.84ARR561 pKa = 11.84LLTQMAQSLAKK572 pKa = 10.03PP573 pKa = 3.71
Molecular weight: 62.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
983 |
410 |
573 |
491.5 |
54.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.664 ± 0.223 | 1.221 ± 0.161 |
4.374 ± 0.011 | 6.307 ± 0.672 |
3.662 ± 0.002 | 7.121 ± 1.492 |
3.052 ± 0.079 | 3.561 ± 0.227 |
5.697 ± 0.267 | 9.461 ± 0.683 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.255 ± 0.543 | 3.255 ± 0.056 |
5.29 ± 0.05 | 2.95 ± 0.471 |
5.799 ± 0.126 | 7.935 ± 1.222 |
6.002 ± 0.585 | 5.697 ± 0.869 |
2.136 ± 1.337 | 3.561 ± 0.714 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
