
Roseicyclus mahoneyensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseicyclus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
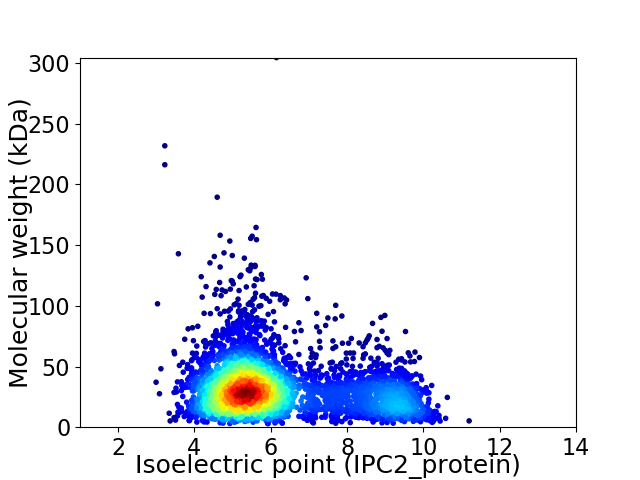
Virtual 2D-PAGE plot for 3941 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
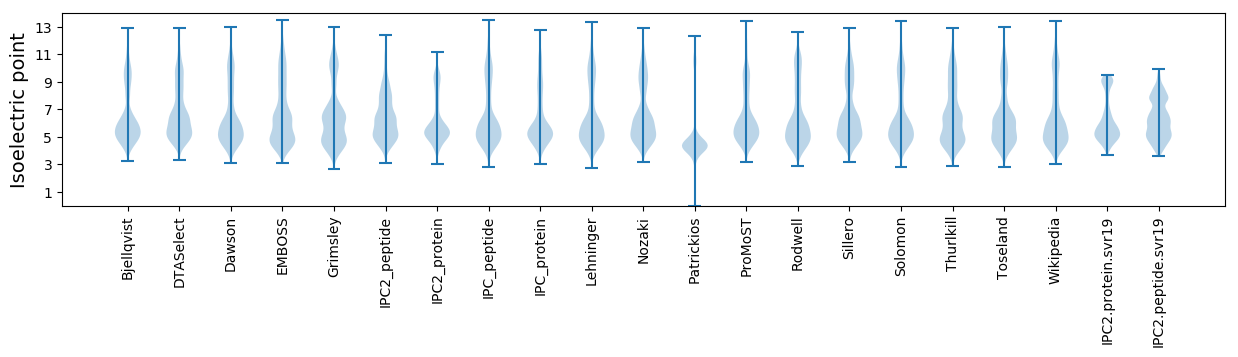
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316GIP0|A0A316GIP0_9RHOB Heterotetrameric sarcosine oxidase alpha subunit OS=Roseicyclus mahoneyensis OX=164332 GN=C7455_103102 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVAFAGAAAADD21 pKa = 3.8VTVTGSAEE29 pKa = 3.7MGIAGGTGLDD39 pKa = 3.28VAFFQSVDD47 pKa = 3.12VRR49 pKa = 11.84FALTGQTDD57 pKa = 3.13NGLSFGATIDD67 pKa = 4.82LDD69 pKa = 4.18DD70 pKa = 5.35ASDD73 pKa = 3.78RR74 pKa = 11.84PLTTGLGDD82 pKa = 3.27ISSAFADD89 pKa = 3.59FTVFVRR95 pKa = 11.84GSFGTLTLGDD105 pKa = 3.59TDD107 pKa = 5.34GAFDD111 pKa = 3.43WAMTEE116 pKa = 4.21VNAGSPGSINDD127 pKa = 3.57AEE129 pKa = 4.46TGHH132 pKa = 6.75GGFNGNSGYY141 pKa = 10.96DD142 pKa = 3.57GDD144 pKa = 4.92HH145 pKa = 6.59NGQILRR151 pKa = 11.84YY152 pKa = 8.92DD153 pKa = 3.61YY154 pKa = 11.02TVGDD158 pKa = 3.76FGFALSAEE166 pKa = 4.43VQGNNPSASANTDD179 pKa = 3.26DD180 pKa = 6.13AIIGLGFRR188 pKa = 11.84YY189 pKa = 10.38GLDD192 pKa = 3.44FGGGRR197 pKa = 11.84LNLGLGYY204 pKa = 8.43QTGDD208 pKa = 2.99HH209 pKa = 6.44GGVGISDD216 pKa = 5.51LIGVSASVALDD227 pKa = 3.3SGLTATVNYY236 pKa = 10.65SEE238 pKa = 5.4GDD240 pKa = 3.5LTLGSLDD247 pKa = 3.57EE248 pKa = 4.16THH250 pKa = 6.88IGLGVSYY257 pKa = 10.16EE258 pKa = 3.9FDD260 pKa = 5.41AITLHH265 pKa = 6.67ANWGQFEE272 pKa = 4.5EE273 pKa = 4.74EE274 pKa = 4.27TTGNTNEE281 pKa = 4.13GFGLAAAYY289 pKa = 10.04DD290 pKa = 4.08LGGGASMHH298 pKa = 6.59LGYY301 pKa = 10.85GDD303 pKa = 4.25SDD305 pKa = 4.23CGGAGPSCTDD315 pKa = 3.26GSRR318 pKa = 11.84WSLGLAMSFF327 pKa = 3.97
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVAFAGAAAADD21 pKa = 3.8VTVTGSAEE29 pKa = 3.7MGIAGGTGLDD39 pKa = 3.28VAFFQSVDD47 pKa = 3.12VRR49 pKa = 11.84FALTGQTDD57 pKa = 3.13NGLSFGATIDD67 pKa = 4.82LDD69 pKa = 4.18DD70 pKa = 5.35ASDD73 pKa = 3.78RR74 pKa = 11.84PLTTGLGDD82 pKa = 3.27ISSAFADD89 pKa = 3.59FTVFVRR95 pKa = 11.84GSFGTLTLGDD105 pKa = 3.59TDD107 pKa = 5.34GAFDD111 pKa = 3.43WAMTEE116 pKa = 4.21VNAGSPGSINDD127 pKa = 3.57AEE129 pKa = 4.46TGHH132 pKa = 6.75GGFNGNSGYY141 pKa = 10.96DD142 pKa = 3.57GDD144 pKa = 4.92HH145 pKa = 6.59NGQILRR151 pKa = 11.84YY152 pKa = 8.92DD153 pKa = 3.61YY154 pKa = 11.02TVGDD158 pKa = 3.76FGFALSAEE166 pKa = 4.43VQGNNPSASANTDD179 pKa = 3.26DD180 pKa = 6.13AIIGLGFRR188 pKa = 11.84YY189 pKa = 10.38GLDD192 pKa = 3.44FGGGRR197 pKa = 11.84LNLGLGYY204 pKa = 8.43QTGDD208 pKa = 2.99HH209 pKa = 6.44GGVGISDD216 pKa = 5.51LIGVSASVALDD227 pKa = 3.3SGLTATVNYY236 pKa = 10.65SEE238 pKa = 5.4GDD240 pKa = 3.5LTLGSLDD247 pKa = 3.57EE248 pKa = 4.16THH250 pKa = 6.88IGLGVSYY257 pKa = 10.16EE258 pKa = 3.9FDD260 pKa = 5.41AITLHH265 pKa = 6.67ANWGQFEE272 pKa = 4.5EE273 pKa = 4.74EE274 pKa = 4.27TTGNTNEE281 pKa = 4.13GFGLAAAYY289 pKa = 10.04DD290 pKa = 4.08LGGGASMHH298 pKa = 6.59LGYY301 pKa = 10.85GDD303 pKa = 4.25SDD305 pKa = 4.23CGGAGPSCTDD315 pKa = 3.26GSRR318 pKa = 11.84WSLGLAMSFF327 pKa = 3.97
Molecular weight: 33.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316G703|A0A316G703_9RHOB ATP-binding cassette subfamily B protein OS=Roseicyclus mahoneyensis OX=164332 GN=C7455_1163 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84AIGRR40 pKa = 11.84KK41 pKa = 9.08SLSAA45 pKa = 3.88
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84AIGRR40 pKa = 11.84KK41 pKa = 9.08SLSAA45 pKa = 3.88
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1221957 |
29 |
2825 |
310.1 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.384 ± 0.063 | 0.849 ± 0.014 |
6.059 ± 0.038 | 5.413 ± 0.04 |
3.59 ± 0.025 | 8.999 ± 0.046 |
2.086 ± 0.023 | 5.033 ± 0.027 |
2.321 ± 0.038 | 10.276 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.768 ± 0.016 | 2.218 ± 0.023 |
5.484 ± 0.034 | 3.055 ± 0.02 |
7.266 ± 0.04 | 4.701 ± 0.027 |
5.642 ± 0.025 | 7.402 ± 0.036 |
1.422 ± 0.018 | 2.031 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
