
Desulfovibrio alaskensis (strain ATCC BAA 1058 / DSM 17464 / G20) (Desulfovibrio desulfuricans (strain G20))
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; Desulfovibrio alaskensis
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
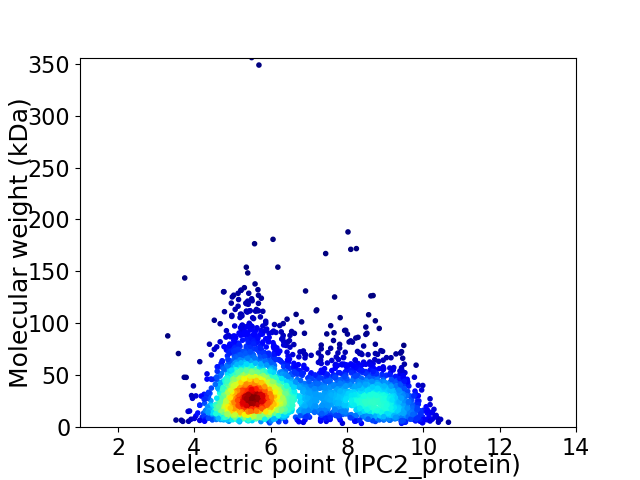
Virtual 2D-PAGE plot for 3220 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
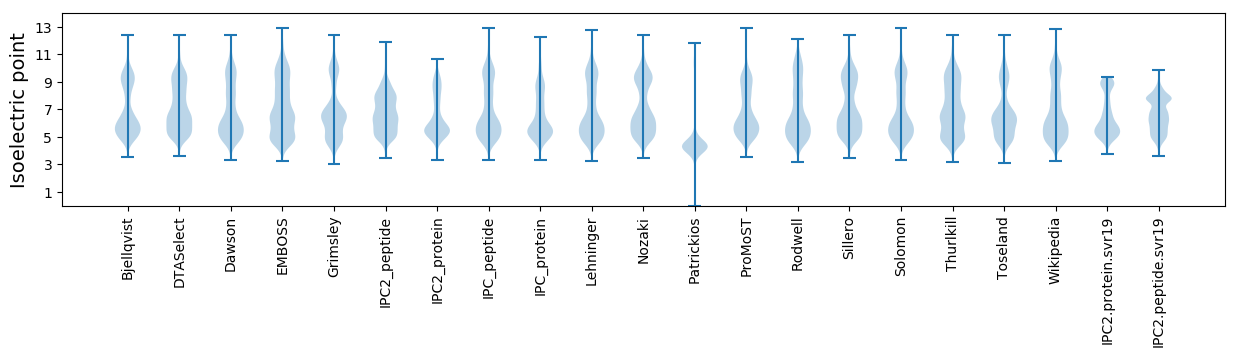
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q30V48|Q30V48_DESAG Aldose 1-epimerase OS=Desulfovibrio alaskensis (strain ATCC BAA 1058 / DSM 17464 / G20) OX=207559 GN=Dde_3655 PE=3 SV=1
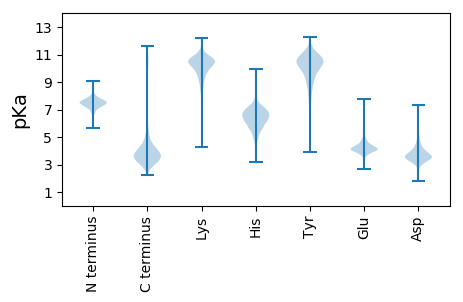
MM1 pKa = 7.51AKK3 pKa = 10.13VLIVYY8 pKa = 9.73GSTTGNTQTTAEE20 pKa = 4.15IVGTEE25 pKa = 4.32LKK27 pKa = 10.81NAGHH31 pKa = 7.09DD32 pKa = 3.44AVVNDD37 pKa = 3.44VTAVEE42 pKa = 4.61AEE44 pKa = 4.47GLCEE48 pKa = 4.97GYY50 pKa = 10.64DD51 pKa = 3.26AVLFGCSTWGDD62 pKa = 3.85EE63 pKa = 4.84EE64 pKa = 6.22IEE66 pKa = 4.17LQDD69 pKa = 5.49DD70 pKa = 5.07FIDD73 pKa = 6.3LYY75 pKa = 11.64DD76 pKa = 4.34EE77 pKa = 4.58LDD79 pKa = 3.39KK80 pKa = 11.74AGLSGVKK87 pKa = 9.75VGCFGCGDD95 pKa = 3.55SSYY98 pKa = 10.9QWFCGAVDD106 pKa = 5.07AIEE109 pKa = 4.28EE110 pKa = 4.11RR111 pKa = 11.84AKK113 pKa = 11.05EE114 pKa = 3.7NGARR118 pKa = 11.84IICDD122 pKa = 3.3SLKK125 pKa = 10.26IDD127 pKa = 4.71GDD129 pKa = 3.9PGSCRR134 pKa = 11.84SEE136 pKa = 3.76IADD139 pKa = 3.38WASAVGAALL148 pKa = 3.81
MM1 pKa = 7.51AKK3 pKa = 10.13VLIVYY8 pKa = 9.73GSTTGNTQTTAEE20 pKa = 4.15IVGTEE25 pKa = 4.32LKK27 pKa = 10.81NAGHH31 pKa = 7.09DD32 pKa = 3.44AVVNDD37 pKa = 3.44VTAVEE42 pKa = 4.61AEE44 pKa = 4.47GLCEE48 pKa = 4.97GYY50 pKa = 10.64DD51 pKa = 3.26AVLFGCSTWGDD62 pKa = 3.85EE63 pKa = 4.84EE64 pKa = 6.22IEE66 pKa = 4.17LQDD69 pKa = 5.49DD70 pKa = 5.07FIDD73 pKa = 6.3LYY75 pKa = 11.64DD76 pKa = 4.34EE77 pKa = 4.58LDD79 pKa = 3.39KK80 pKa = 11.74AGLSGVKK87 pKa = 9.75VGCFGCGDD95 pKa = 3.55SSYY98 pKa = 10.9QWFCGAVDD106 pKa = 5.07AIEE109 pKa = 4.28EE110 pKa = 4.11RR111 pKa = 11.84AKK113 pKa = 11.05EE114 pKa = 3.7NGARR118 pKa = 11.84IICDD122 pKa = 3.3SLKK125 pKa = 10.26IDD127 pKa = 4.71GDD129 pKa = 3.9PGSCRR134 pKa = 11.84SEE136 pKa = 3.76IADD139 pKa = 3.38WASAVGAALL148 pKa = 3.81
Molecular weight: 15.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q315Z7|Q315Z7_DESAG ABC-type transporter periplasmic subunit OS=Desulfovibrio alaskensis (strain ATCC BAA 1058 / DSM 17464 / G20) OX=207559 GN=Dde_0448 PE=4 SV=1
MM1 pKa = 8.02KK2 pKa = 8.89YY3 pKa = 7.73TCTITRR9 pKa = 11.84HH10 pKa = 4.93SSRR13 pKa = 11.84GTLRR17 pKa = 11.84GKK19 pKa = 8.55TVLRR23 pKa = 11.84AAPGEE28 pKa = 4.39VSWHH32 pKa = 5.87IALKK36 pKa = 9.84ILGYY40 pKa = 10.21ILFMPHH46 pKa = 5.55TPRR49 pKa = 11.84IEE51 pKa = 4.39EE52 pKa = 3.82NVGWHH57 pKa = 6.4FKK59 pKa = 10.34PDD61 pKa = 3.46LAAYY65 pKa = 9.64APDD68 pKa = 4.04GGVSLWVDD76 pKa = 3.91CGNIAVRR83 pKa = 11.84KK84 pKa = 8.18ISRR87 pKa = 11.84VAAWLPAAAEE97 pKa = 3.75FHH99 pKa = 6.0ILRR102 pKa = 11.84RR103 pKa = 11.84TVRR106 pKa = 11.84DD107 pKa = 3.29ASLLCASARR116 pKa = 11.84AVRR119 pKa = 11.84RR120 pKa = 11.84PEE122 pKa = 3.92RR123 pKa = 11.84VHH125 pKa = 6.1ITAFDD130 pKa = 3.36NGFVHH135 pKa = 6.74TLADD139 pKa = 3.65MLDD142 pKa = 3.25ATNRR146 pKa = 11.84MEE148 pKa = 5.22AVLDD152 pKa = 3.63NHH154 pKa = 6.75RR155 pKa = 11.84LALTVTCRR163 pKa = 11.84SGTATLHH170 pKa = 5.85SALHH174 pKa = 6.51RR175 pKa = 11.84LTPASGNPARR185 pKa = 11.84PRR187 pKa = 11.84SS188 pKa = 3.62
MM1 pKa = 8.02KK2 pKa = 8.89YY3 pKa = 7.73TCTITRR9 pKa = 11.84HH10 pKa = 4.93SSRR13 pKa = 11.84GTLRR17 pKa = 11.84GKK19 pKa = 8.55TVLRR23 pKa = 11.84AAPGEE28 pKa = 4.39VSWHH32 pKa = 5.87IALKK36 pKa = 9.84ILGYY40 pKa = 10.21ILFMPHH46 pKa = 5.55TPRR49 pKa = 11.84IEE51 pKa = 4.39EE52 pKa = 3.82NVGWHH57 pKa = 6.4FKK59 pKa = 10.34PDD61 pKa = 3.46LAAYY65 pKa = 9.64APDD68 pKa = 4.04GGVSLWVDD76 pKa = 3.91CGNIAVRR83 pKa = 11.84KK84 pKa = 8.18ISRR87 pKa = 11.84VAAWLPAAAEE97 pKa = 3.75FHH99 pKa = 6.0ILRR102 pKa = 11.84RR103 pKa = 11.84TVRR106 pKa = 11.84DD107 pKa = 3.29ASLLCASARR116 pKa = 11.84AVRR119 pKa = 11.84RR120 pKa = 11.84PEE122 pKa = 3.92RR123 pKa = 11.84VHH125 pKa = 6.1ITAFDD130 pKa = 3.36NGFVHH135 pKa = 6.74TLADD139 pKa = 3.65MLDD142 pKa = 3.25ATNRR146 pKa = 11.84MEE148 pKa = 5.22AVLDD152 pKa = 3.63NHH154 pKa = 6.75RR155 pKa = 11.84LALTVTCRR163 pKa = 11.84SGTATLHH170 pKa = 5.85SALHH174 pKa = 6.51RR175 pKa = 11.84LTPASGNPARR185 pKa = 11.84PRR187 pKa = 11.84SS188 pKa = 3.62
Molecular weight: 20.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1069418 |
31 |
3252 |
332.1 |
36.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.903 ± 0.061 | 1.452 ± 0.022 |
5.378 ± 0.033 | 6.132 ± 0.038 |
3.776 ± 0.028 | 8.033 ± 0.032 |
2.345 ± 0.023 | 4.812 ± 0.037 |
3.713 ± 0.037 | 10.349 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.893 ± 0.022 | 2.805 ± 0.022 |
4.811 ± 0.029 | 3.619 ± 0.026 |
6.926 ± 0.038 | 5.49 ± 0.03 |
5.308 ± 0.029 | 7.489 ± 0.04 |
1.179 ± 0.018 | 2.588 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
