
Devosia sp. YR412
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Devosia; unclassified Devosia
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
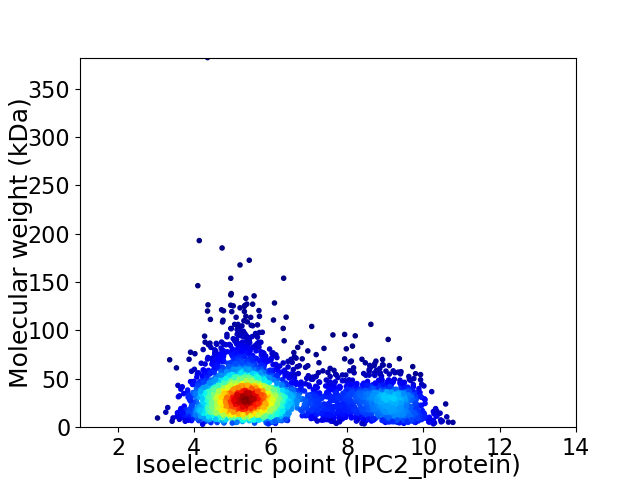
Virtual 2D-PAGE plot for 3685 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8ZY17|A0A1H8ZY17_9RHIZ Iron complex transport system ATP-binding protein OS=Devosia sp. YR412 OX=1881030 GN=SAMN05428969_0494 PE=3 SV=1
MM1 pKa = 7.94LDD3 pKa = 3.53GLTLGEE9 pKa = 3.55IHH11 pKa = 6.33MRR13 pKa = 11.84LVLVIALLLSTAPAFAADD31 pKa = 3.38AVIYY35 pKa = 10.56KK36 pKa = 8.49GTLGGKK42 pKa = 9.97DD43 pKa = 2.86IVVEE47 pKa = 4.12LTDD50 pKa = 3.92PSADD54 pKa = 3.27QVVGRR59 pKa = 11.84YY60 pKa = 9.31SYY62 pKa = 11.2LSQGGDD68 pKa = 3.08IPLSSVDD75 pKa = 3.28TSEE78 pKa = 4.83GVVTLAEE85 pKa = 4.15EE86 pKa = 4.71APCTEE91 pKa = 4.37TTCVFDD97 pKa = 4.65DD98 pKa = 3.76NYY100 pKa = 10.8EE101 pKa = 4.06IVEE104 pKa = 4.33VPIGAHH110 pKa = 3.88WQLRR114 pKa = 11.84VKK116 pKa = 10.72DD117 pKa = 4.44DD118 pKa = 3.71GSITGGWNPADD129 pKa = 3.58GSAVLDD135 pKa = 3.58IEE137 pKa = 4.29LTEE140 pKa = 4.12VARR143 pKa = 11.84RR144 pKa = 11.84TLPEE148 pKa = 4.54DD149 pKa = 3.5MAITPADD156 pKa = 3.84LADD159 pKa = 4.1SAWVASYY166 pKa = 11.08DD167 pKa = 3.57NPAAFSRR174 pKa = 11.84DD175 pKa = 3.13SAPYY179 pKa = 10.36DD180 pKa = 3.53FAKK183 pKa = 10.09MDD185 pKa = 3.46VALTEE190 pKa = 4.58GPTRR194 pKa = 11.84TMDD197 pKa = 3.34GNSYY201 pKa = 10.76RR202 pKa = 11.84DD203 pKa = 3.68VIDD206 pKa = 4.23PRR208 pKa = 11.84SKK210 pKa = 10.23FPFPRR215 pKa = 11.84VVAFADD221 pKa = 4.57GSPVEE226 pKa = 4.18AANTRR231 pKa = 11.84LAARR235 pKa = 11.84HH236 pKa = 5.26AQINMSAFACLSKK249 pKa = 10.76AYY251 pKa = 10.18AGLGYY256 pKa = 9.7RR257 pKa = 11.84DD258 pKa = 3.46GMGGGGLGDD267 pKa = 4.04FDD269 pKa = 4.76GEE271 pKa = 4.59SIEE274 pKa = 4.33IAYY277 pKa = 9.82LSPTVINWTEE287 pKa = 3.93TGSTYY292 pKa = 10.73CGGAYY297 pKa = 9.43PNNHH301 pKa = 6.43FDD303 pKa = 4.53SYY305 pKa = 11.1IIDD308 pKa = 3.72ARR310 pKa = 11.84TGADD314 pKa = 3.84FALGWVLKK322 pKa = 10.48DD323 pKa = 3.85WIATADD329 pKa = 3.52TTNYY333 pKa = 10.52EE334 pKa = 4.15PVTDD338 pKa = 3.69QAAALEE344 pKa = 4.38NPNGYY349 pKa = 9.69LFAPGQPLIDD359 pKa = 3.7YY360 pKa = 10.15ALAYY364 pKa = 10.34LDD366 pKa = 4.43EE367 pKa = 6.24AGLDD371 pKa = 3.4PDD373 pKa = 5.18LASEE377 pKa = 4.39CQLPDD382 pKa = 4.89LIAEE386 pKa = 4.51GNLGFRR392 pKa = 11.84FAPEE396 pKa = 3.79NKK398 pKa = 9.76LVFAITGLAHH408 pKa = 7.27ANFACSEE415 pKa = 4.17DD416 pKa = 3.83VLTVSLSDD424 pKa = 3.49IPEE427 pKa = 4.35LLALTASDD435 pKa = 4.48YY436 pKa = 11.27FPDD439 pKa = 4.14LANN442 pKa = 3.68
MM1 pKa = 7.94LDD3 pKa = 3.53GLTLGEE9 pKa = 3.55IHH11 pKa = 6.33MRR13 pKa = 11.84LVLVIALLLSTAPAFAADD31 pKa = 3.38AVIYY35 pKa = 10.56KK36 pKa = 8.49GTLGGKK42 pKa = 9.97DD43 pKa = 2.86IVVEE47 pKa = 4.12LTDD50 pKa = 3.92PSADD54 pKa = 3.27QVVGRR59 pKa = 11.84YY60 pKa = 9.31SYY62 pKa = 11.2LSQGGDD68 pKa = 3.08IPLSSVDD75 pKa = 3.28TSEE78 pKa = 4.83GVVTLAEE85 pKa = 4.15EE86 pKa = 4.71APCTEE91 pKa = 4.37TTCVFDD97 pKa = 4.65DD98 pKa = 3.76NYY100 pKa = 10.8EE101 pKa = 4.06IVEE104 pKa = 4.33VPIGAHH110 pKa = 3.88WQLRR114 pKa = 11.84VKK116 pKa = 10.72DD117 pKa = 4.44DD118 pKa = 3.71GSITGGWNPADD129 pKa = 3.58GSAVLDD135 pKa = 3.58IEE137 pKa = 4.29LTEE140 pKa = 4.12VARR143 pKa = 11.84RR144 pKa = 11.84TLPEE148 pKa = 4.54DD149 pKa = 3.5MAITPADD156 pKa = 3.84LADD159 pKa = 4.1SAWVASYY166 pKa = 11.08DD167 pKa = 3.57NPAAFSRR174 pKa = 11.84DD175 pKa = 3.13SAPYY179 pKa = 10.36DD180 pKa = 3.53FAKK183 pKa = 10.09MDD185 pKa = 3.46VALTEE190 pKa = 4.58GPTRR194 pKa = 11.84TMDD197 pKa = 3.34GNSYY201 pKa = 10.76RR202 pKa = 11.84DD203 pKa = 3.68VIDD206 pKa = 4.23PRR208 pKa = 11.84SKK210 pKa = 10.23FPFPRR215 pKa = 11.84VVAFADD221 pKa = 4.57GSPVEE226 pKa = 4.18AANTRR231 pKa = 11.84LAARR235 pKa = 11.84HH236 pKa = 5.26AQINMSAFACLSKK249 pKa = 10.76AYY251 pKa = 10.18AGLGYY256 pKa = 9.7RR257 pKa = 11.84DD258 pKa = 3.46GMGGGGLGDD267 pKa = 4.04FDD269 pKa = 4.76GEE271 pKa = 4.59SIEE274 pKa = 4.33IAYY277 pKa = 9.82LSPTVINWTEE287 pKa = 3.93TGSTYY292 pKa = 10.73CGGAYY297 pKa = 9.43PNNHH301 pKa = 6.43FDD303 pKa = 4.53SYY305 pKa = 11.1IIDD308 pKa = 3.72ARR310 pKa = 11.84TGADD314 pKa = 3.84FALGWVLKK322 pKa = 10.48DD323 pKa = 3.85WIATADD329 pKa = 3.52TTNYY333 pKa = 10.52EE334 pKa = 4.15PVTDD338 pKa = 3.69QAAALEE344 pKa = 4.38NPNGYY349 pKa = 9.69LFAPGQPLIDD359 pKa = 3.7YY360 pKa = 10.15ALAYY364 pKa = 10.34LDD366 pKa = 4.43EE367 pKa = 6.24AGLDD371 pKa = 3.4PDD373 pKa = 5.18LASEE377 pKa = 4.39CQLPDD382 pKa = 4.89LIAEE386 pKa = 4.51GNLGFRR392 pKa = 11.84FAPEE396 pKa = 3.79NKK398 pKa = 9.76LVFAITGLAHH408 pKa = 7.27ANFACSEE415 pKa = 4.17DD416 pKa = 3.83VLTVSLSDD424 pKa = 3.49IPEE427 pKa = 4.35LLALTASDD435 pKa = 4.48YY436 pKa = 11.27FPDD439 pKa = 4.14LANN442 pKa = 3.68
Molecular weight: 47.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9CDL2|A0A1H9CDL2_9RHIZ Transcriptional regulator HxlR family OS=Devosia sp. YR412 OX=1881030 GN=SAMN05428969_1445 PE=4 SV=1
MM1 pKa = 7.67PALILGLAVLLVVTMSVAVGAGAVPIPVDD30 pKa = 3.75TVWRR34 pKa = 11.84IIASKK39 pKa = 9.89LAPGSVVADD48 pKa = 3.43WTSGRR53 pKa = 11.84EE54 pKa = 3.93NIVWEE59 pKa = 4.15VRR61 pKa = 11.84LPRR64 pKa = 11.84IVLGAIVGASLAVVGAALQSVTRR87 pKa = 11.84NPLADD92 pKa = 3.56PHH94 pKa = 7.59LLGISSGAAFGAILVLLHH112 pKa = 6.37TGMFLGLITVPLFAFIGALIATALVLAVSNLTKK145 pKa = 10.48SQSAGRR151 pKa = 11.84LVLAGVAVSFIITAAGNLFIFLGDD175 pKa = 3.23PRR177 pKa = 11.84AAHH180 pKa = 5.55TVVFWMLGGLGLAQWQQLPFPLAALVLCGGYY211 pKa = 10.48LILKK215 pKa = 9.56ARR217 pKa = 11.84SLNAMTLGDD226 pKa = 4.11EE227 pKa = 4.45SATTLGIPALRR238 pKa = 11.84FRR240 pKa = 11.84LTVFVICALLTGSAVAFSGVIAFVGLMIPHH270 pKa = 7.6IVRR273 pKa = 11.84MMVGGDD279 pKa = 3.5YY280 pKa = 10.81RR281 pKa = 11.84RR282 pKa = 11.84VLPLSALVGAIFLVLADD299 pKa = 3.81IAARR303 pKa = 11.84TLMPPQDD310 pKa = 3.76MPIGVLTGIIGGLFFVGLMRR330 pKa = 11.84WRR332 pKa = 11.84KK333 pKa = 9.87SGEE336 pKa = 3.7
MM1 pKa = 7.67PALILGLAVLLVVTMSVAVGAGAVPIPVDD30 pKa = 3.75TVWRR34 pKa = 11.84IIASKK39 pKa = 9.89LAPGSVVADD48 pKa = 3.43WTSGRR53 pKa = 11.84EE54 pKa = 3.93NIVWEE59 pKa = 4.15VRR61 pKa = 11.84LPRR64 pKa = 11.84IVLGAIVGASLAVVGAALQSVTRR87 pKa = 11.84NPLADD92 pKa = 3.56PHH94 pKa = 7.59LLGISSGAAFGAILVLLHH112 pKa = 6.37TGMFLGLITVPLFAFIGALIATALVLAVSNLTKK145 pKa = 10.48SQSAGRR151 pKa = 11.84LVLAGVAVSFIITAAGNLFIFLGDD175 pKa = 3.23PRR177 pKa = 11.84AAHH180 pKa = 5.55TVVFWMLGGLGLAQWQQLPFPLAALVLCGGYY211 pKa = 10.48LILKK215 pKa = 9.56ARR217 pKa = 11.84SLNAMTLGDD226 pKa = 4.11EE227 pKa = 4.45SATTLGIPALRR238 pKa = 11.84FRR240 pKa = 11.84LTVFVICALLTGSAVAFSGVIAFVGLMIPHH270 pKa = 7.6IVRR273 pKa = 11.84MMVGGDD279 pKa = 3.5YY280 pKa = 10.81RR281 pKa = 11.84RR282 pKa = 11.84VLPLSALVGAIFLVLADD299 pKa = 3.81IAARR303 pKa = 11.84TLMPPQDD310 pKa = 3.76MPIGVLTGIIGGLFFVGLMRR330 pKa = 11.84WRR332 pKa = 11.84KK333 pKa = 9.87SGEE336 pKa = 3.7
Molecular weight: 34.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1143512 |
29 |
3895 |
310.3 |
33.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.346 ± 0.058 | 0.627 ± 0.01 |
5.772 ± 0.032 | 5.517 ± 0.034 |
3.805 ± 0.027 | 8.47 ± 0.039 |
1.952 ± 0.019 | 5.52 ± 0.026 |
3.288 ± 0.034 | 10.323 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.022 | 2.862 ± 0.027 |
4.942 ± 0.029 | 3.327 ± 0.026 |
6.122 ± 0.038 | 5.566 ± 0.03 |
5.708 ± 0.045 | 7.695 ± 0.033 |
1.303 ± 0.019 | 2.286 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
