
Alkalihalobacillus nanhaiisediminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
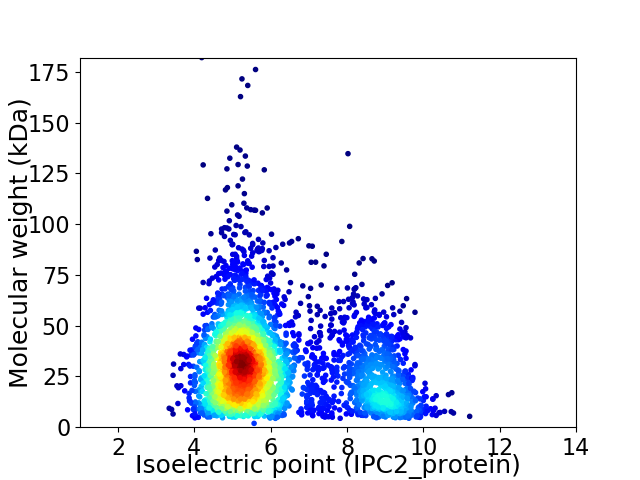
Virtual 2D-PAGE plot for 3479 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562QSY2|A0A562QSY2_9BACI Probable lipid II flippase MurJ OS=Alkalihalobacillus nanhaiisediminis OX=688079 GN=murJ PE=3 SV=1
MM1 pKa = 7.23KK2 pKa = 10.26KK3 pKa = 9.84WMISLGLALVLTACGGSEE21 pKa = 4.13TEE23 pKa = 4.57EE24 pKa = 4.21TTEE27 pKa = 4.12TPAEE31 pKa = 4.18GSEE34 pKa = 4.1EE35 pKa = 3.92AVEE38 pKa = 4.16EE39 pKa = 4.73GSDD42 pKa = 3.65EE43 pKa = 4.15VFEE46 pKa = 4.3IAVIPSQSIGEE57 pKa = 4.32MEE59 pKa = 3.97TGLNNLEE66 pKa = 3.77EE67 pKa = 4.37HH68 pKa = 6.36LADD71 pKa = 3.68RR72 pKa = 11.84LGRR75 pKa = 11.84EE76 pKa = 4.42VVVEE80 pKa = 3.96QYY82 pKa = 10.93PNYY85 pKa = 9.52NAVVEE90 pKa = 4.19ALNYY94 pKa = 10.19NHH96 pKa = 7.49IDD98 pKa = 3.57LAYY101 pKa = 10.28LGPLTYY107 pKa = 10.64LIAHH111 pKa = 6.27EE112 pKa = 4.08QSGAKK117 pKa = 10.14AIITQEE123 pKa = 3.84IDD125 pKa = 3.04GSPYY129 pKa = 10.01YY130 pKa = 10.3YY131 pKa = 11.14SNIITHH137 pKa = 7.37ADD139 pKa = 3.58AEE141 pKa = 4.49WEE143 pKa = 4.04NLEE146 pKa = 5.83DD147 pKa = 3.75MLEE150 pKa = 4.24DD151 pKa = 3.43RR152 pKa = 11.84AEE154 pKa = 3.87VDD156 pKa = 3.97FAFASISSTSGHH168 pKa = 7.68LIPGLEE174 pKa = 3.97LRR176 pKa = 11.84NLGYY180 pKa = 10.57YY181 pKa = 9.9EE182 pKa = 5.37SEE184 pKa = 4.55DD185 pKa = 3.17NHH187 pKa = 6.41EE188 pKa = 4.27FNQIQFAGSHH198 pKa = 6.2DD199 pKa = 4.0VVATLVQDD207 pKa = 3.53QTVDD211 pKa = 2.92AGAIDD216 pKa = 3.99SAILEE221 pKa = 4.22ALMTEE226 pKa = 4.52DD227 pKa = 4.13EE228 pKa = 4.68NNGGTLRR235 pKa = 11.84DD236 pKa = 4.4DD237 pKa = 4.73IKK239 pKa = 11.36VIWQSEE245 pKa = 3.94QLYY248 pKa = 9.13QYY250 pKa = 9.25PWVVPAQMDD259 pKa = 3.88DD260 pKa = 3.55EE261 pKa = 5.78LISQVQEE268 pKa = 3.53AFYY271 pKa = 10.38EE272 pKa = 4.26IEE274 pKa = 4.29DD275 pKa = 3.97EE276 pKa = 4.49EE277 pKa = 4.22ILRR280 pKa = 11.84IFGGATKK287 pKa = 10.1FVEE290 pKa = 4.91ADD292 pKa = 3.22DD293 pKa = 4.06SQYY296 pKa = 11.89ADD298 pKa = 3.24VLEE301 pKa = 4.44AARR304 pKa = 11.84EE305 pKa = 3.83FDD307 pKa = 3.46MLTLEE312 pKa = 4.5EE313 pKa = 4.23EE314 pKa = 4.36
MM1 pKa = 7.23KK2 pKa = 10.26KK3 pKa = 9.84WMISLGLALVLTACGGSEE21 pKa = 4.13TEE23 pKa = 4.57EE24 pKa = 4.21TTEE27 pKa = 4.12TPAEE31 pKa = 4.18GSEE34 pKa = 4.1EE35 pKa = 3.92AVEE38 pKa = 4.16EE39 pKa = 4.73GSDD42 pKa = 3.65EE43 pKa = 4.15VFEE46 pKa = 4.3IAVIPSQSIGEE57 pKa = 4.32MEE59 pKa = 3.97TGLNNLEE66 pKa = 3.77EE67 pKa = 4.37HH68 pKa = 6.36LADD71 pKa = 3.68RR72 pKa = 11.84LGRR75 pKa = 11.84EE76 pKa = 4.42VVVEE80 pKa = 3.96QYY82 pKa = 10.93PNYY85 pKa = 9.52NAVVEE90 pKa = 4.19ALNYY94 pKa = 10.19NHH96 pKa = 7.49IDD98 pKa = 3.57LAYY101 pKa = 10.28LGPLTYY107 pKa = 10.64LIAHH111 pKa = 6.27EE112 pKa = 4.08QSGAKK117 pKa = 10.14AIITQEE123 pKa = 3.84IDD125 pKa = 3.04GSPYY129 pKa = 10.01YY130 pKa = 10.3YY131 pKa = 11.14SNIITHH137 pKa = 7.37ADD139 pKa = 3.58AEE141 pKa = 4.49WEE143 pKa = 4.04NLEE146 pKa = 5.83DD147 pKa = 3.75MLEE150 pKa = 4.24DD151 pKa = 3.43RR152 pKa = 11.84AEE154 pKa = 3.87VDD156 pKa = 3.97FAFASISSTSGHH168 pKa = 7.68LIPGLEE174 pKa = 3.97LRR176 pKa = 11.84NLGYY180 pKa = 10.57YY181 pKa = 9.9EE182 pKa = 5.37SEE184 pKa = 4.55DD185 pKa = 3.17NHH187 pKa = 6.41EE188 pKa = 4.27FNQIQFAGSHH198 pKa = 6.2DD199 pKa = 4.0VVATLVQDD207 pKa = 3.53QTVDD211 pKa = 2.92AGAIDD216 pKa = 3.99SAILEE221 pKa = 4.22ALMTEE226 pKa = 4.52DD227 pKa = 4.13EE228 pKa = 4.68NNGGTLRR235 pKa = 11.84DD236 pKa = 4.4DD237 pKa = 4.73IKK239 pKa = 11.36VIWQSEE245 pKa = 3.94QLYY248 pKa = 9.13QYY250 pKa = 9.25PWVVPAQMDD259 pKa = 3.88DD260 pKa = 3.55EE261 pKa = 5.78LISQVQEE268 pKa = 3.53AFYY271 pKa = 10.38EE272 pKa = 4.26IEE274 pKa = 4.29DD275 pKa = 3.97EE276 pKa = 4.49EE277 pKa = 4.22ILRR280 pKa = 11.84IFGGATKK287 pKa = 10.1FVEE290 pKa = 4.91ADD292 pKa = 3.22DD293 pKa = 4.06SQYY296 pKa = 11.89ADD298 pKa = 3.24VLEE301 pKa = 4.44AARR304 pKa = 11.84EE305 pKa = 3.83FDD307 pKa = 3.46MLTLEE312 pKa = 4.5EE313 pKa = 4.23EE314 pKa = 4.36
Molecular weight: 35.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562QSG2|A0A562QSG2_9BACI Uncharacterized protein OS=Alkalihalobacillus nanhaiisediminis OX=688079 GN=IQ10_00832 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.15KK15 pKa = 9.35KK16 pKa = 9.06HH17 pKa = 4.22GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.22NGRR29 pKa = 11.84QVLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.15KK15 pKa = 9.35KK16 pKa = 9.06HH17 pKa = 4.22GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.22NGRR29 pKa = 11.84QVLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1004815 |
16 |
1686 |
288.8 |
32.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.239 ± 0.045 | 0.7 ± 0.011 |
5.053 ± 0.03 | 7.895 ± 0.051 |
4.36 ± 0.032 | 6.866 ± 0.041 |
2.213 ± 0.022 | 7.7 ± 0.038 |
6.303 ± 0.038 | 9.821 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.849 ± 0.019 | 4.057 ± 0.03 |
3.601 ± 0.023 | 4.048 ± 0.033 |
4.366 ± 0.028 | 5.843 ± 0.031 |
5.51 ± 0.027 | 7.315 ± 0.032 |
0.967 ± 0.016 | 3.295 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
