
Lymantria dispar cypovirus 1 (isolate Rao) (LdCPV-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; Cypovirus 1; Lymantria dispar cypovirus 1
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
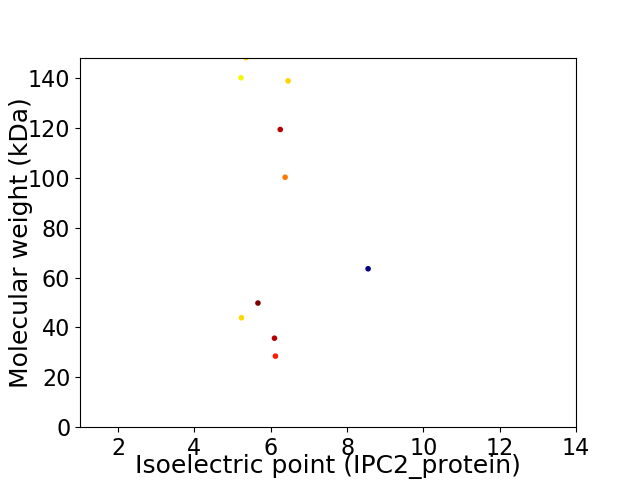
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>sp|Q91IE0|RDRP_LDCPR RNA-directed RNA polymerase VP2 OS=Lymantria dispar cypovirus 1 (isolate Rao) OX=648169 GN=S2 PE=3 SV=1
MM1 pKa = 7.8EE2 pKa = 5.25INRR5 pKa = 11.84AEE7 pKa = 3.92IRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.4ITRR14 pKa = 11.84YY15 pKa = 10.16AGLIEE20 pKa = 4.38QQTQINITDD29 pKa = 3.6NDD31 pKa = 3.28QDD33 pKa = 3.6ILKK36 pKa = 9.07TLIADD41 pKa = 3.38YY42 pKa = 10.11NLRR45 pKa = 11.84MRR47 pKa = 11.84RR48 pKa = 11.84DD49 pKa = 3.38ALLGEE54 pKa = 4.63LARR57 pKa = 11.84LDD59 pKa = 3.87EE60 pKa = 5.09LRR62 pKa = 11.84DD63 pKa = 3.31ISQIKK68 pKa = 9.22GDD70 pKa = 4.15EE71 pKa = 4.21YY72 pKa = 11.58KK73 pKa = 10.23LTIPLLPIISTLNQHH88 pKa = 5.82EE89 pKa = 4.37FEE91 pKa = 5.45IIQANIEE98 pKa = 3.95TDD100 pKa = 4.76FIADD104 pKa = 3.29NVTFITSFIPADD116 pKa = 3.9LDD118 pKa = 3.76LEE120 pKa = 4.3QTIQHH125 pKa = 5.1VFFRR129 pKa = 11.84TTATTPYY136 pKa = 9.3FRR138 pKa = 11.84SFNLVIAILDD148 pKa = 3.69YY149 pKa = 11.68DD150 pKa = 4.12EE151 pKa = 6.21DD152 pKa = 4.54KK153 pKa = 11.7GDD155 pKa = 3.28VKK157 pKa = 11.25LDD159 pKa = 3.41VKK161 pKa = 10.29ITITRR166 pKa = 11.84SNNGVFNYY174 pKa = 10.46NYY176 pKa = 8.37TWAGKK181 pKa = 9.9DD182 pKa = 3.44YY183 pKa = 10.43EE184 pKa = 4.53RR185 pKa = 11.84VSICYY190 pKa = 10.26NLISYY195 pKa = 7.52LQQINGPRR203 pKa = 11.84GRR205 pKa = 11.84DD206 pKa = 3.52DD207 pKa = 3.57EE208 pKa = 6.21AEE210 pKa = 3.99MPIYY214 pKa = 10.31EE215 pKa = 4.87IVRR218 pKa = 11.84QNNGSQPSYY227 pKa = 11.28ASGEE231 pKa = 3.89HH232 pKa = 6.21LYY234 pKa = 10.25IVSSHH239 pKa = 5.54LHH241 pKa = 5.12VDD243 pKa = 3.98EE244 pKa = 4.28IVRR247 pKa = 11.84DD248 pKa = 3.84RR249 pKa = 11.84EE250 pKa = 4.07HH251 pKa = 7.41RR252 pKa = 11.84DD253 pKa = 2.75ISVDD257 pKa = 3.54VTEE260 pKa = 5.09LNLMFPIVRR269 pKa = 11.84MFDD272 pKa = 3.47PVDD275 pKa = 3.57LRR277 pKa = 11.84DD278 pKa = 3.54IRR280 pKa = 11.84IEE282 pKa = 3.99DD283 pKa = 3.56VTPGIEE289 pKa = 3.88FTINMEE295 pKa = 3.67VSTYY299 pKa = 8.92LTEE302 pKa = 4.47LSGSHH307 pKa = 6.81VDD309 pKa = 3.56TQRR312 pKa = 11.84TIMNHH317 pKa = 4.95AEE319 pKa = 4.24KK320 pKa = 10.39IVGNYY325 pKa = 7.86TGQQWNVQSNMLSEE339 pKa = 4.28VRR341 pKa = 11.84TQKK344 pKa = 10.97LEE346 pKa = 4.13EE347 pKa = 4.2EE348 pKa = 4.48DD349 pKa = 3.46EE350 pKa = 4.37EE351 pKa = 4.13ARR353 pKa = 11.84QRR355 pKa = 11.84GDD357 pKa = 3.31YY358 pKa = 7.49TTSTLVQTMAQVSDD372 pKa = 4.38LFSSTILYY380 pKa = 10.0RR381 pKa = 11.84YY382 pKa = 10.3AEE384 pKa = 4.57AEE386 pKa = 3.88LDD388 pKa = 3.67NTVGAFEE395 pKa = 4.56LLRR398 pKa = 11.84PVMSIPTEE406 pKa = 4.17YY407 pKa = 10.24IHH409 pKa = 7.45DD410 pKa = 3.89GRR412 pKa = 11.84IGPITNISASASIVTSSNNGVGEE435 pKa = 3.82VRR437 pKa = 11.84NIFKK441 pKa = 10.47PIGDD445 pKa = 3.53QTINEE450 pKa = 3.85AHH452 pKa = 6.5FANVYY457 pKa = 11.05SNDD460 pKa = 2.94EE461 pKa = 3.84YY462 pKa = 11.44AIYY465 pKa = 10.73LRR467 pKa = 11.84FSYY470 pKa = 10.17RR471 pKa = 11.84QAPVQSEE478 pKa = 4.27TVYY481 pKa = 10.68LQQALPSMRR490 pKa = 11.84IVSPSSVSTTVSTALIGGNTIRR512 pKa = 11.84INCPIRR518 pKa = 11.84PHH520 pKa = 7.86RR521 pKa = 11.84EE522 pKa = 3.81DD523 pKa = 3.49NFVAGGVQIPRR534 pKa = 11.84QSTAVEE540 pKa = 3.65IHH542 pKa = 5.29VQEE545 pKa = 3.89ILIGYY550 pKa = 7.75RR551 pKa = 11.84QATTFPIDD559 pKa = 3.34TEE561 pKa = 4.46GRR563 pKa = 11.84LSLEE567 pKa = 3.75LMYY570 pKa = 11.09GLEE573 pKa = 4.17SRR575 pKa = 11.84SAVGNTMSPVRR586 pKa = 11.84FVTVNDD592 pKa = 4.14GEE594 pKa = 4.67FFGLTCPIDD603 pKa = 3.72LTLSTIVDD611 pKa = 3.7PASYY615 pKa = 10.61LSDD618 pKa = 3.27GVILVTTAFEE628 pKa = 4.37DD629 pKa = 3.53LRR631 pKa = 11.84GYY633 pKa = 10.96AWLATLGGDD642 pKa = 3.65WPRR645 pKa = 11.84TYY647 pKa = 10.57NSSMGAFNIFTGGDD661 pKa = 3.17INLSTEE667 pKa = 4.12YY668 pKa = 10.32GSEE671 pKa = 3.52MTYY674 pKa = 9.04TFKK677 pKa = 11.33VEE679 pKa = 4.22LPINYY684 pKa = 7.66MFNNMTISSHH694 pKa = 4.72NVPRR698 pKa = 11.84VPVLGVTYY706 pKa = 10.82ASIYY710 pKa = 9.37QDD712 pKa = 2.74SRR714 pKa = 11.84TDD716 pKa = 3.34LEE718 pKa = 4.03ARR720 pKa = 11.84RR721 pKa = 11.84FLQTLVFRR729 pKa = 11.84IHH731 pKa = 6.74GSWSARR737 pKa = 11.84VPYY740 pKa = 10.16PPGNLPTRR748 pKa = 11.84NTANQHH754 pKa = 4.71QDD756 pKa = 2.53IQQVINDD763 pKa = 4.73SIFQEE768 pKa = 4.21LDD770 pKa = 3.13RR771 pKa = 11.84LSDD774 pKa = 3.64EE775 pKa = 5.56LLDD778 pKa = 3.84LEE780 pKa = 4.86NRR782 pKa = 11.84LDD784 pKa = 3.46HH785 pKa = 7.34LEE787 pKa = 4.05RR788 pKa = 11.84QFEE791 pKa = 4.23MFIQSQEE798 pKa = 4.27SEE800 pKa = 3.45WWEE803 pKa = 3.49ILLNVVIDD811 pKa = 3.36ISIGYY816 pKa = 9.52FSTFAGDD823 pKa = 3.63ALKK826 pKa = 10.59NAQRR830 pKa = 11.84AITKK834 pKa = 9.92AVGYY838 pKa = 6.45TRR840 pKa = 11.84RR841 pKa = 11.84VLMTVTKK848 pKa = 9.57TMRR851 pKa = 11.84NGTIFTRR858 pKa = 11.84LLGAKK863 pKa = 9.24NLSGQALASLEE874 pKa = 4.2TLVEE878 pKa = 4.07SALRR882 pKa = 11.84SINMKK887 pKa = 9.71KK888 pKa = 10.1SRR890 pKa = 11.84FMRR893 pKa = 11.84GAEE896 pKa = 3.93PLYY899 pKa = 9.56KK900 pKa = 9.53TNKK903 pKa = 7.95VAQHH907 pKa = 6.45IDD909 pKa = 3.25NTEE912 pKa = 3.93KK913 pKa = 10.4MNMMMDD919 pKa = 3.49FSFANRR925 pKa = 11.84NNRR928 pKa = 11.84QNITADD934 pKa = 3.82TLSKK938 pKa = 9.8MHH940 pKa = 6.22TQNAHH945 pKa = 5.01GTSDD949 pKa = 3.83TILPAMRR956 pKa = 11.84VYY958 pKa = 10.47YY959 pKa = 10.39RR960 pKa = 11.84PLGFLDD966 pKa = 3.67KK967 pKa = 10.76RR968 pKa = 11.84VGDD971 pKa = 4.09ALHH974 pKa = 6.36TGITRR979 pKa = 11.84PEE981 pKa = 3.84ALKK984 pKa = 10.49KK985 pKa = 9.73QLRR988 pKa = 11.84SDD990 pKa = 3.66VANVGTRR997 pKa = 11.84APSHH1001 pKa = 6.6AFMTYY1006 pKa = 9.71TDD1008 pKa = 3.94VLYY1011 pKa = 10.46EE1012 pKa = 4.15DD1013 pKa = 3.98AGSYY1017 pKa = 9.77IVSKK1021 pKa = 10.3RR1022 pKa = 11.84YY1023 pKa = 10.23LGIGEE1028 pKa = 4.33LNKK1031 pKa = 10.33FGRR1034 pKa = 11.84TTSDD1038 pKa = 2.71KK1039 pKa = 10.31NAGIGGVNIKK1049 pKa = 10.52YY1050 pKa = 9.9RR1051 pKa = 11.84VNKK1054 pKa = 8.24ITADD1058 pKa = 3.03GKK1060 pKa = 10.68YY1061 pKa = 10.19IIDD1064 pKa = 3.54RR1065 pKa = 11.84LDD1067 pKa = 3.55YY1068 pKa = 10.73TEE1070 pKa = 4.69SGYY1073 pKa = 8.57TALDD1077 pKa = 3.2VDD1079 pKa = 4.52RR1080 pKa = 11.84LYY1082 pKa = 11.55SSLFGKK1088 pKa = 10.28QGDD1091 pKa = 4.18GLSTEE1096 pKa = 4.43QKK1098 pKa = 10.11WMDD1101 pKa = 2.94ISKK1104 pKa = 10.91GVDD1107 pKa = 2.97AKK1109 pKa = 10.74IISADD1114 pKa = 3.47MVSEE1118 pKa = 4.26EE1119 pKa = 4.38FLSSKK1124 pKa = 9.54YY1125 pKa = 8.25TGQMIDD1131 pKa = 3.87EE1132 pKa = 5.93LINSPPQFNYY1142 pKa = 10.36SLVYY1146 pKa = 10.52RR1147 pKa = 11.84NCQDD1151 pKa = 2.99FALDD1155 pKa = 3.65VLRR1158 pKa = 11.84VAQGFSPSNKK1168 pKa = 8.11WDD1170 pKa = 3.19VSTAARR1176 pKa = 11.84MQQRR1180 pKa = 11.84RR1181 pKa = 11.84VISLMDD1187 pKa = 4.15DD1188 pKa = 3.4LMGEE1192 pKa = 4.29SEE1194 pKa = 4.19TFARR1198 pKa = 11.84SGRR1201 pKa = 11.84ASQLLLRR1208 pKa = 11.84QVRR1211 pKa = 11.84EE1212 pKa = 4.25SYY1214 pKa = 10.64VKK1216 pKa = 10.06ARR1218 pKa = 11.84KK1219 pKa = 9.85RR1220 pKa = 11.84GDD1222 pKa = 3.41LQAVKK1227 pKa = 10.48ALQLRR1232 pKa = 11.84FKK1234 pKa = 11.33GFFF1237 pKa = 3.29
MM1 pKa = 7.8EE2 pKa = 5.25INRR5 pKa = 11.84AEE7 pKa = 3.92IRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.4ITRR14 pKa = 11.84YY15 pKa = 10.16AGLIEE20 pKa = 4.38QQTQINITDD29 pKa = 3.6NDD31 pKa = 3.28QDD33 pKa = 3.6ILKK36 pKa = 9.07TLIADD41 pKa = 3.38YY42 pKa = 10.11NLRR45 pKa = 11.84MRR47 pKa = 11.84RR48 pKa = 11.84DD49 pKa = 3.38ALLGEE54 pKa = 4.63LARR57 pKa = 11.84LDD59 pKa = 3.87EE60 pKa = 5.09LRR62 pKa = 11.84DD63 pKa = 3.31ISQIKK68 pKa = 9.22GDD70 pKa = 4.15EE71 pKa = 4.21YY72 pKa = 11.58KK73 pKa = 10.23LTIPLLPIISTLNQHH88 pKa = 5.82EE89 pKa = 4.37FEE91 pKa = 5.45IIQANIEE98 pKa = 3.95TDD100 pKa = 4.76FIADD104 pKa = 3.29NVTFITSFIPADD116 pKa = 3.9LDD118 pKa = 3.76LEE120 pKa = 4.3QTIQHH125 pKa = 5.1VFFRR129 pKa = 11.84TTATTPYY136 pKa = 9.3FRR138 pKa = 11.84SFNLVIAILDD148 pKa = 3.69YY149 pKa = 11.68DD150 pKa = 4.12EE151 pKa = 6.21DD152 pKa = 4.54KK153 pKa = 11.7GDD155 pKa = 3.28VKK157 pKa = 11.25LDD159 pKa = 3.41VKK161 pKa = 10.29ITITRR166 pKa = 11.84SNNGVFNYY174 pKa = 10.46NYY176 pKa = 8.37TWAGKK181 pKa = 9.9DD182 pKa = 3.44YY183 pKa = 10.43EE184 pKa = 4.53RR185 pKa = 11.84VSICYY190 pKa = 10.26NLISYY195 pKa = 7.52LQQINGPRR203 pKa = 11.84GRR205 pKa = 11.84DD206 pKa = 3.52DD207 pKa = 3.57EE208 pKa = 6.21AEE210 pKa = 3.99MPIYY214 pKa = 10.31EE215 pKa = 4.87IVRR218 pKa = 11.84QNNGSQPSYY227 pKa = 11.28ASGEE231 pKa = 3.89HH232 pKa = 6.21LYY234 pKa = 10.25IVSSHH239 pKa = 5.54LHH241 pKa = 5.12VDD243 pKa = 3.98EE244 pKa = 4.28IVRR247 pKa = 11.84DD248 pKa = 3.84RR249 pKa = 11.84EE250 pKa = 4.07HH251 pKa = 7.41RR252 pKa = 11.84DD253 pKa = 2.75ISVDD257 pKa = 3.54VTEE260 pKa = 5.09LNLMFPIVRR269 pKa = 11.84MFDD272 pKa = 3.47PVDD275 pKa = 3.57LRR277 pKa = 11.84DD278 pKa = 3.54IRR280 pKa = 11.84IEE282 pKa = 3.99DD283 pKa = 3.56VTPGIEE289 pKa = 3.88FTINMEE295 pKa = 3.67VSTYY299 pKa = 8.92LTEE302 pKa = 4.47LSGSHH307 pKa = 6.81VDD309 pKa = 3.56TQRR312 pKa = 11.84TIMNHH317 pKa = 4.95AEE319 pKa = 4.24KK320 pKa = 10.39IVGNYY325 pKa = 7.86TGQQWNVQSNMLSEE339 pKa = 4.28VRR341 pKa = 11.84TQKK344 pKa = 10.97LEE346 pKa = 4.13EE347 pKa = 4.2EE348 pKa = 4.48DD349 pKa = 3.46EE350 pKa = 4.37EE351 pKa = 4.13ARR353 pKa = 11.84QRR355 pKa = 11.84GDD357 pKa = 3.31YY358 pKa = 7.49TTSTLVQTMAQVSDD372 pKa = 4.38LFSSTILYY380 pKa = 10.0RR381 pKa = 11.84YY382 pKa = 10.3AEE384 pKa = 4.57AEE386 pKa = 3.88LDD388 pKa = 3.67NTVGAFEE395 pKa = 4.56LLRR398 pKa = 11.84PVMSIPTEE406 pKa = 4.17YY407 pKa = 10.24IHH409 pKa = 7.45DD410 pKa = 3.89GRR412 pKa = 11.84IGPITNISASASIVTSSNNGVGEE435 pKa = 3.82VRR437 pKa = 11.84NIFKK441 pKa = 10.47PIGDD445 pKa = 3.53QTINEE450 pKa = 3.85AHH452 pKa = 6.5FANVYY457 pKa = 11.05SNDD460 pKa = 2.94EE461 pKa = 3.84YY462 pKa = 11.44AIYY465 pKa = 10.73LRR467 pKa = 11.84FSYY470 pKa = 10.17RR471 pKa = 11.84QAPVQSEE478 pKa = 4.27TVYY481 pKa = 10.68LQQALPSMRR490 pKa = 11.84IVSPSSVSTTVSTALIGGNTIRR512 pKa = 11.84INCPIRR518 pKa = 11.84PHH520 pKa = 7.86RR521 pKa = 11.84EE522 pKa = 3.81DD523 pKa = 3.49NFVAGGVQIPRR534 pKa = 11.84QSTAVEE540 pKa = 3.65IHH542 pKa = 5.29VQEE545 pKa = 3.89ILIGYY550 pKa = 7.75RR551 pKa = 11.84QATTFPIDD559 pKa = 3.34TEE561 pKa = 4.46GRR563 pKa = 11.84LSLEE567 pKa = 3.75LMYY570 pKa = 11.09GLEE573 pKa = 4.17SRR575 pKa = 11.84SAVGNTMSPVRR586 pKa = 11.84FVTVNDD592 pKa = 4.14GEE594 pKa = 4.67FFGLTCPIDD603 pKa = 3.72LTLSTIVDD611 pKa = 3.7PASYY615 pKa = 10.61LSDD618 pKa = 3.27GVILVTTAFEE628 pKa = 4.37DD629 pKa = 3.53LRR631 pKa = 11.84GYY633 pKa = 10.96AWLATLGGDD642 pKa = 3.65WPRR645 pKa = 11.84TYY647 pKa = 10.57NSSMGAFNIFTGGDD661 pKa = 3.17INLSTEE667 pKa = 4.12YY668 pKa = 10.32GSEE671 pKa = 3.52MTYY674 pKa = 9.04TFKK677 pKa = 11.33VEE679 pKa = 4.22LPINYY684 pKa = 7.66MFNNMTISSHH694 pKa = 4.72NVPRR698 pKa = 11.84VPVLGVTYY706 pKa = 10.82ASIYY710 pKa = 9.37QDD712 pKa = 2.74SRR714 pKa = 11.84TDD716 pKa = 3.34LEE718 pKa = 4.03ARR720 pKa = 11.84RR721 pKa = 11.84FLQTLVFRR729 pKa = 11.84IHH731 pKa = 6.74GSWSARR737 pKa = 11.84VPYY740 pKa = 10.16PPGNLPTRR748 pKa = 11.84NTANQHH754 pKa = 4.71QDD756 pKa = 2.53IQQVINDD763 pKa = 4.73SIFQEE768 pKa = 4.21LDD770 pKa = 3.13RR771 pKa = 11.84LSDD774 pKa = 3.64EE775 pKa = 5.56LLDD778 pKa = 3.84LEE780 pKa = 4.86NRR782 pKa = 11.84LDD784 pKa = 3.46HH785 pKa = 7.34LEE787 pKa = 4.05RR788 pKa = 11.84QFEE791 pKa = 4.23MFIQSQEE798 pKa = 4.27SEE800 pKa = 3.45WWEE803 pKa = 3.49ILLNVVIDD811 pKa = 3.36ISIGYY816 pKa = 9.52FSTFAGDD823 pKa = 3.63ALKK826 pKa = 10.59NAQRR830 pKa = 11.84AITKK834 pKa = 9.92AVGYY838 pKa = 6.45TRR840 pKa = 11.84RR841 pKa = 11.84VLMTVTKK848 pKa = 9.57TMRR851 pKa = 11.84NGTIFTRR858 pKa = 11.84LLGAKK863 pKa = 9.24NLSGQALASLEE874 pKa = 4.2TLVEE878 pKa = 4.07SALRR882 pKa = 11.84SINMKK887 pKa = 9.71KK888 pKa = 10.1SRR890 pKa = 11.84FMRR893 pKa = 11.84GAEE896 pKa = 3.93PLYY899 pKa = 9.56KK900 pKa = 9.53TNKK903 pKa = 7.95VAQHH907 pKa = 6.45IDD909 pKa = 3.25NTEE912 pKa = 3.93KK913 pKa = 10.4MNMMMDD919 pKa = 3.49FSFANRR925 pKa = 11.84NNRR928 pKa = 11.84QNITADD934 pKa = 3.82TLSKK938 pKa = 9.8MHH940 pKa = 6.22TQNAHH945 pKa = 5.01GTSDD949 pKa = 3.83TILPAMRR956 pKa = 11.84VYY958 pKa = 10.47YY959 pKa = 10.39RR960 pKa = 11.84PLGFLDD966 pKa = 3.67KK967 pKa = 10.76RR968 pKa = 11.84VGDD971 pKa = 4.09ALHH974 pKa = 6.36TGITRR979 pKa = 11.84PEE981 pKa = 3.84ALKK984 pKa = 10.49KK985 pKa = 9.73QLRR988 pKa = 11.84SDD990 pKa = 3.66VANVGTRR997 pKa = 11.84APSHH1001 pKa = 6.6AFMTYY1006 pKa = 9.71TDD1008 pKa = 3.94VLYY1011 pKa = 10.46EE1012 pKa = 4.15DD1013 pKa = 3.98AGSYY1017 pKa = 9.77IVSKK1021 pKa = 10.3RR1022 pKa = 11.84YY1023 pKa = 10.23LGIGEE1028 pKa = 4.33LNKK1031 pKa = 10.33FGRR1034 pKa = 11.84TTSDD1038 pKa = 2.71KK1039 pKa = 10.31NAGIGGVNIKK1049 pKa = 10.52YY1050 pKa = 9.9RR1051 pKa = 11.84VNKK1054 pKa = 8.24ITADD1058 pKa = 3.03GKK1060 pKa = 10.68YY1061 pKa = 10.19IIDD1064 pKa = 3.54RR1065 pKa = 11.84LDD1067 pKa = 3.55YY1068 pKa = 10.73TEE1070 pKa = 4.69SGYY1073 pKa = 8.57TALDD1077 pKa = 3.2VDD1079 pKa = 4.52RR1080 pKa = 11.84LYY1082 pKa = 11.55SSLFGKK1088 pKa = 10.28QGDD1091 pKa = 4.18GLSTEE1096 pKa = 4.43QKK1098 pKa = 10.11WMDD1101 pKa = 2.94ISKK1104 pKa = 10.91GVDD1107 pKa = 2.97AKK1109 pKa = 10.74IISADD1114 pKa = 3.47MVSEE1118 pKa = 4.26EE1119 pKa = 4.38FLSSKK1124 pKa = 9.54YY1125 pKa = 8.25TGQMIDD1131 pKa = 3.87EE1132 pKa = 5.93LINSPPQFNYY1142 pKa = 10.36SLVYY1146 pKa = 10.52RR1147 pKa = 11.84NCQDD1151 pKa = 2.99FALDD1155 pKa = 3.65VLRR1158 pKa = 11.84VAQGFSPSNKK1168 pKa = 8.11WDD1170 pKa = 3.19VSTAARR1176 pKa = 11.84MQQRR1180 pKa = 11.84RR1181 pKa = 11.84VISLMDD1187 pKa = 4.15DD1188 pKa = 3.4LMGEE1192 pKa = 4.29SEE1194 pKa = 4.19TFARR1198 pKa = 11.84SGRR1201 pKa = 11.84ASQLLLRR1208 pKa = 11.84QVRR1211 pKa = 11.84EE1212 pKa = 4.25SYY1214 pKa = 10.64VKK1216 pKa = 10.06ARR1218 pKa = 11.84KK1219 pKa = 9.85RR1220 pKa = 11.84GDD1222 pKa = 3.41LQAVKK1227 pKa = 10.48ALQLRR1232 pKa = 11.84FKK1234 pKa = 11.33GFFF1237 pKa = 3.29
Molecular weight: 140.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91ID7|NS1_LDCPR Non-structural protein 1 OS=Lymantria dispar cypovirus 1 (isolate Rao) OX=648169 GN=S5 PE=4 SV=1
MM1 pKa = 7.39FAIDD5 pKa = 4.05PLKK8 pKa = 10.53HH9 pKa = 5.65SKK11 pKa = 10.57LYY13 pKa = 10.59EE14 pKa = 4.03EE15 pKa = 4.63YY16 pKa = 10.94GLYY19 pKa = 10.45LRR21 pKa = 11.84PHH23 pKa = 6.9QINQEE28 pKa = 3.99IKK30 pKa = 8.54PTTIKK35 pKa = 10.54KK36 pKa = 10.23KK37 pKa = 10.17EE38 pKa = 3.85LAPTIRR44 pKa = 11.84SIRR47 pKa = 11.84YY48 pKa = 8.68ASLLHH53 pKa = 7.24SMLAKK58 pKa = 10.14HH59 pKa = 6.53AARR62 pKa = 11.84HH63 pKa = 5.68NGTLINPRR71 pKa = 11.84MYY73 pKa = 11.37ADD75 pKa = 5.11MITLGNTKK83 pKa = 8.74VTVTKK88 pKa = 8.63GTPKK92 pKa = 10.7AQIDD96 pKa = 3.92TLKK99 pKa = 10.59MNGLTVVSKK108 pKa = 10.56SRR110 pKa = 11.84RR111 pKa = 11.84NNKK114 pKa = 9.1KK115 pKa = 10.34KK116 pKa = 10.17PVSDD120 pKa = 3.6TTATTDD126 pKa = 3.13EE127 pKa = 4.27NTNDD131 pKa = 2.75IVTYY135 pKa = 10.27KK136 pKa = 10.86ALTEE140 pKa = 3.87MSTLVEE146 pKa = 4.44SFHH149 pKa = 6.77LPSGLTLIVFDD160 pKa = 4.26DD161 pKa = 4.14EE162 pKa = 5.54KK163 pKa = 10.49YY164 pKa = 10.26QSLIPDD170 pKa = 4.67YY171 pKa = 10.49INQLITYY178 pKa = 6.7TQPHH182 pKa = 7.76IIPTWQGIADD192 pKa = 4.33FSDD195 pKa = 4.24PYY197 pKa = 10.45LRR199 pKa = 11.84SYY201 pKa = 10.55FRR203 pKa = 11.84RR204 pKa = 11.84PFEE207 pKa = 3.98LTASNLASPQKK218 pKa = 10.69HH219 pKa = 5.52NLSPLTRR226 pKa = 11.84SIFNNTGRR234 pKa = 11.84EE235 pKa = 3.67DD236 pKa = 4.54AIIKK240 pKa = 9.84KK241 pKa = 10.17LYY243 pKa = 10.62GYY245 pKa = 10.49GEE247 pKa = 4.57YY248 pKa = 10.27IFIKK252 pKa = 10.95YY253 pKa = 8.52EE254 pKa = 3.93GCLITWTGVYY264 pKa = 10.03GAVAMMVNLPKK275 pKa = 9.92RR276 pKa = 11.84DD277 pKa = 3.76LGLDD281 pKa = 3.29AGDD284 pKa = 4.3NFLKK288 pKa = 10.35EE289 pKa = 4.2YY290 pKa = 10.32KK291 pKa = 9.94QLLFYY296 pKa = 10.92GVITDD301 pKa = 4.31ATPSGISAKK310 pKa = 9.49STVIKK315 pKa = 10.61ISPHH319 pKa = 7.29KK320 pKa = 10.05MMNPSGGALAVLSKK334 pKa = 10.58FLEE337 pKa = 4.46AVVSANVINATLVVYY352 pKa = 10.51AEE354 pKa = 4.34KK355 pKa = 10.85GAGKK359 pKa = 8.68TSFLSTYY366 pKa = 10.83AEE368 pKa = 4.07RR369 pKa = 11.84LSVASGQAVGHH380 pKa = 6.87LSSDD384 pKa = 3.19AYY386 pKa = 10.65GRR388 pKa = 11.84WLAKK392 pKa = 10.62NKK394 pKa = 9.73DD395 pKa = 3.49VKK397 pKa = 10.78EE398 pKa = 3.97PSFEE402 pKa = 3.88YY403 pKa = 10.43DD404 pKa = 3.37YY405 pKa = 11.71VLSRR409 pKa = 11.84DD410 pKa = 3.47TDD412 pKa = 3.7DD413 pKa = 3.54TEE415 pKa = 4.58SYY417 pKa = 9.97YY418 pKa = 9.3EE419 pKa = 3.89QRR421 pKa = 11.84ASEE424 pKa = 4.55LLTLHH429 pKa = 7.08GISEE433 pKa = 4.5LAQYY437 pKa = 10.37EE438 pKa = 4.3LLSVRR443 pKa = 11.84KK444 pKa = 8.78KK445 pKa = 9.85VKK447 pKa = 9.58MMNEE451 pKa = 3.71MNEE454 pKa = 3.77ILIAQLEE461 pKa = 4.32NADD464 pKa = 3.67THH466 pKa = 6.13SEE468 pKa = 4.04RR469 pKa = 11.84NFYY472 pKa = 11.58YY473 pKa = 9.99MVSTGKK479 pKa = 8.32NTPRR483 pKa = 11.84TLIVEE488 pKa = 4.11GHH490 pKa = 6.37FNAQDD495 pKa = 3.26ATIARR500 pKa = 11.84TDD502 pKa = 3.41TTVLLRR508 pKa = 11.84TINDD512 pKa = 3.35TTQAMRR518 pKa = 11.84DD519 pKa = 3.56RR520 pKa = 11.84QRR522 pKa = 11.84SGVVQLFLRR531 pKa = 11.84DD532 pKa = 3.14TYY534 pKa = 11.62YY535 pKa = 11.33RR536 pKa = 11.84LLPSLHH542 pKa = 4.97TTVYY546 pKa = 8.82PFEE549 pKa = 4.09MLEE552 pKa = 4.54SIKK555 pKa = 10.15RR556 pKa = 11.84WKK558 pKa = 8.44WVHH561 pKa = 5.43
MM1 pKa = 7.39FAIDD5 pKa = 4.05PLKK8 pKa = 10.53HH9 pKa = 5.65SKK11 pKa = 10.57LYY13 pKa = 10.59EE14 pKa = 4.03EE15 pKa = 4.63YY16 pKa = 10.94GLYY19 pKa = 10.45LRR21 pKa = 11.84PHH23 pKa = 6.9QINQEE28 pKa = 3.99IKK30 pKa = 8.54PTTIKK35 pKa = 10.54KK36 pKa = 10.23KK37 pKa = 10.17EE38 pKa = 3.85LAPTIRR44 pKa = 11.84SIRR47 pKa = 11.84YY48 pKa = 8.68ASLLHH53 pKa = 7.24SMLAKK58 pKa = 10.14HH59 pKa = 6.53AARR62 pKa = 11.84HH63 pKa = 5.68NGTLINPRR71 pKa = 11.84MYY73 pKa = 11.37ADD75 pKa = 5.11MITLGNTKK83 pKa = 8.74VTVTKK88 pKa = 8.63GTPKK92 pKa = 10.7AQIDD96 pKa = 3.92TLKK99 pKa = 10.59MNGLTVVSKK108 pKa = 10.56SRR110 pKa = 11.84RR111 pKa = 11.84NNKK114 pKa = 9.1KK115 pKa = 10.34KK116 pKa = 10.17PVSDD120 pKa = 3.6TTATTDD126 pKa = 3.13EE127 pKa = 4.27NTNDD131 pKa = 2.75IVTYY135 pKa = 10.27KK136 pKa = 10.86ALTEE140 pKa = 3.87MSTLVEE146 pKa = 4.44SFHH149 pKa = 6.77LPSGLTLIVFDD160 pKa = 4.26DD161 pKa = 4.14EE162 pKa = 5.54KK163 pKa = 10.49YY164 pKa = 10.26QSLIPDD170 pKa = 4.67YY171 pKa = 10.49INQLITYY178 pKa = 6.7TQPHH182 pKa = 7.76IIPTWQGIADD192 pKa = 4.33FSDD195 pKa = 4.24PYY197 pKa = 10.45LRR199 pKa = 11.84SYY201 pKa = 10.55FRR203 pKa = 11.84RR204 pKa = 11.84PFEE207 pKa = 3.98LTASNLASPQKK218 pKa = 10.69HH219 pKa = 5.52NLSPLTRR226 pKa = 11.84SIFNNTGRR234 pKa = 11.84EE235 pKa = 3.67DD236 pKa = 4.54AIIKK240 pKa = 9.84KK241 pKa = 10.17LYY243 pKa = 10.62GYY245 pKa = 10.49GEE247 pKa = 4.57YY248 pKa = 10.27IFIKK252 pKa = 10.95YY253 pKa = 8.52EE254 pKa = 3.93GCLITWTGVYY264 pKa = 10.03GAVAMMVNLPKK275 pKa = 9.92RR276 pKa = 11.84DD277 pKa = 3.76LGLDD281 pKa = 3.29AGDD284 pKa = 4.3NFLKK288 pKa = 10.35EE289 pKa = 4.2YY290 pKa = 10.32KK291 pKa = 9.94QLLFYY296 pKa = 10.92GVITDD301 pKa = 4.31ATPSGISAKK310 pKa = 9.49STVIKK315 pKa = 10.61ISPHH319 pKa = 7.29KK320 pKa = 10.05MMNPSGGALAVLSKK334 pKa = 10.58FLEE337 pKa = 4.46AVVSANVINATLVVYY352 pKa = 10.51AEE354 pKa = 4.34KK355 pKa = 10.85GAGKK359 pKa = 8.68TSFLSTYY366 pKa = 10.83AEE368 pKa = 4.07RR369 pKa = 11.84LSVASGQAVGHH380 pKa = 6.87LSSDD384 pKa = 3.19AYY386 pKa = 10.65GRR388 pKa = 11.84WLAKK392 pKa = 10.62NKK394 pKa = 9.73DD395 pKa = 3.49VKK397 pKa = 10.78EE398 pKa = 3.97PSFEE402 pKa = 3.88YY403 pKa = 10.43DD404 pKa = 3.37YY405 pKa = 11.71VLSRR409 pKa = 11.84DD410 pKa = 3.47TDD412 pKa = 3.7DD413 pKa = 3.54TEE415 pKa = 4.58SYY417 pKa = 9.97YY418 pKa = 9.3EE419 pKa = 3.89QRR421 pKa = 11.84ASEE424 pKa = 4.55LLTLHH429 pKa = 7.08GISEE433 pKa = 4.5LAQYY437 pKa = 10.37EE438 pKa = 4.3LLSVRR443 pKa = 11.84KK444 pKa = 8.78KK445 pKa = 9.85VKK447 pKa = 9.58MMNEE451 pKa = 3.71MNEE454 pKa = 3.77ILIAQLEE461 pKa = 4.32NADD464 pKa = 3.67THH466 pKa = 6.13SEE468 pKa = 4.04RR469 pKa = 11.84NFYY472 pKa = 11.58YY473 pKa = 9.99MVSTGKK479 pKa = 8.32NTPRR483 pKa = 11.84TLIVEE488 pKa = 4.11GHH490 pKa = 6.37FNAQDD495 pKa = 3.26ATIARR500 pKa = 11.84TDD502 pKa = 3.41TTVLLRR508 pKa = 11.84TINDD512 pKa = 3.35TTQAMRR518 pKa = 11.84DD519 pKa = 3.56RR520 pKa = 11.84QRR522 pKa = 11.84SGVVQLFLRR531 pKa = 11.84DD532 pKa = 3.14TYY534 pKa = 11.62YY535 pKa = 11.33RR536 pKa = 11.84LLPSLHH542 pKa = 4.97TTVYY546 pKa = 8.82PFEE549 pKa = 4.09MLEE552 pKa = 4.54SIKK555 pKa = 10.15RR556 pKa = 11.84WKK558 pKa = 8.44WVHH561 pKa = 5.43
Molecular weight: 63.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7701 |
248 |
1333 |
770.1 |
86.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.324 ± 0.478 | 0.727 ± 0.184 |
5.752 ± 0.252 | 5.61 ± 0.305 |
4.012 ± 0.249 | 4.869 ± 0.274 |
2.558 ± 0.195 | 6.739 ± 0.383 |
4.441 ± 0.575 | 8.752 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.701 ± 0.172 | 5.506 ± 0.275 |
4.207 ± 0.3 | 3.818 ± 0.224 |
5.843 ± 0.316 | 7.869 ± 0.26 |
7.596 ± 0.415 | 6.584 ± 0.244 |
0.766 ± 0.077 | 4.324 ± 0.307 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
