
Tamlana fucoidanivorans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tamlana
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
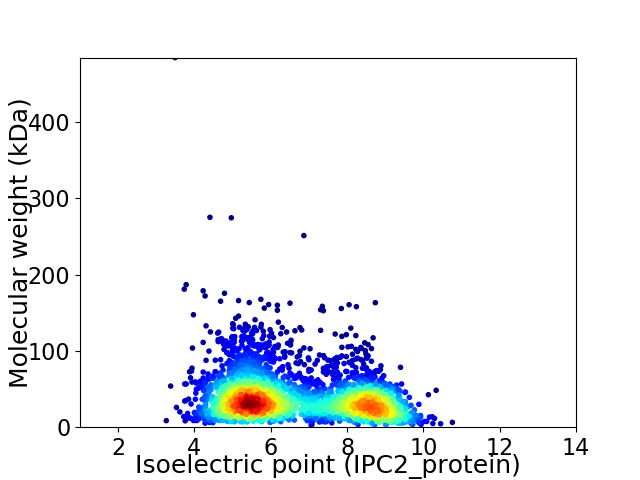
Virtual 2D-PAGE plot for 3334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C4SP99|A0A5C4SP99_9FLAO (2R 3S)-2-methylisocitrate dehydratase OS=Tamlana fucoidanivorans OX=2583814 GN=FGF67_04835 PE=4 SV=1
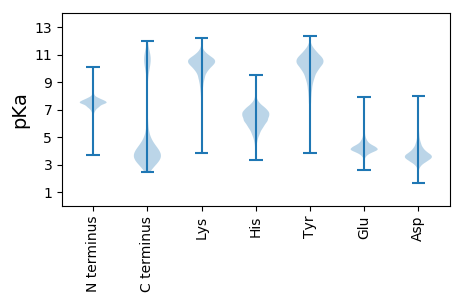
MM1 pKa = 7.47KK2 pKa = 10.24KK3 pKa = 9.74ILNICMMVITIIALNSCEE21 pKa = 4.35EE22 pKa = 4.21KK23 pKa = 10.23DD24 pKa = 3.4TSEE27 pKa = 5.74FIAQEE32 pKa = 3.85ATDD35 pKa = 4.16FTFSTSFLEE44 pKa = 5.21NYY46 pKa = 9.92LLNNSLGNNLAEE58 pKa = 4.13RR59 pKa = 11.84FTWRR63 pKa = 11.84DD64 pKa = 3.05ANFGIPTEE72 pKa = 4.16RR73 pKa = 11.84TYY75 pKa = 11.31EE76 pKa = 3.95LQAATMEE83 pKa = 4.68DD84 pKa = 3.62FSDD87 pKa = 3.92YY88 pKa = 11.44TNDD91 pKa = 4.48PDD93 pKa = 4.25DD94 pKa = 4.93SNYY97 pKa = 10.5KK98 pKa = 10.38AYY100 pKa = 11.06NLGTNIGGNEE110 pKa = 3.48IPATVKK116 pKa = 10.5QFLALAKK123 pKa = 10.22IAGLDD128 pKa = 3.72NDD130 pKa = 5.1PEE132 pKa = 4.53TPAPNTGKK140 pKa = 10.75LYY142 pKa = 10.33FRR144 pKa = 11.84VRR146 pKa = 11.84ATIGKK151 pKa = 9.68DD152 pKa = 3.05NLEE155 pKa = 4.24SFSAIQALNIEE166 pKa = 4.26LQEE169 pKa = 4.39GEE171 pKa = 4.55EE172 pKa = 4.25EE173 pKa = 4.38VGGPNCPSWWVVGAGAPDD191 pKa = 4.91AGWNWDD197 pKa = 3.46NPIEE201 pKa = 4.39FLCTDD206 pKa = 3.84QIYY209 pKa = 9.17TSNINLANDD218 pKa = 3.44SFRR221 pKa = 11.84FFTEE225 pKa = 3.29KK226 pKa = 11.07DD227 pKa = 3.12NWDD230 pKa = 3.48SGLNFPYY237 pKa = 10.21FVDD240 pKa = 3.24EE241 pKa = 5.55GYY243 pKa = 10.93SIDD246 pKa = 3.94PNFEE250 pKa = 3.63NAMDD254 pKa = 4.06GDD256 pKa = 4.07EE257 pKa = 4.2NFRR260 pKa = 11.84FIGTPGKK267 pKa = 10.7YY268 pKa = 9.95SLTIDD273 pKa = 4.22SVNKK277 pKa = 9.1TIKK280 pKa = 10.28LGPPAAEE287 pKa = 5.14DD288 pKa = 4.16PNCDD292 pKa = 3.75SVWVVGAGAVDD303 pKa = 5.03AGWNWDD309 pKa = 3.45SPIEE313 pKa = 4.12FFCTDD318 pKa = 2.9NVYY321 pKa = 9.3STNIKK326 pKa = 8.59LTNDD330 pKa = 2.84AFRR333 pKa = 11.84FFLEE337 pKa = 4.0EE338 pKa = 4.39GNWDD342 pKa = 3.38SGQNYY347 pKa = 8.66PFYY350 pKa = 11.24ANDD353 pKa = 4.49GYY355 pKa = 9.72TIDD358 pKa = 5.08SLLEE362 pKa = 4.09DD363 pKa = 4.72AQDD366 pKa = 3.5GDD368 pKa = 4.45NNFKK372 pKa = 11.15FNGTPGTYY380 pKa = 10.55LLTVDD385 pKa = 4.58TVNKK389 pKa = 9.4TITLGIPRR397 pKa = 11.84PEE399 pKa = 4.34EE400 pKa = 4.03PNCDD404 pKa = 3.93SVWVVGAGAVDD415 pKa = 5.03AGWNWDD421 pKa = 3.49SPIEE425 pKa = 4.14FTCTDD430 pKa = 3.32NVYY433 pKa = 10.48SASINLTNDD442 pKa = 2.6AFRR445 pKa = 11.84FFLEE449 pKa = 4.0EE450 pKa = 4.39GNWDD454 pKa = 3.38SGQNYY459 pKa = 8.63PFYY462 pKa = 11.07ADD464 pKa = 4.17DD465 pKa = 5.48GYY467 pKa = 9.64TIDD470 pKa = 4.64TLLEE474 pKa = 4.25DD475 pKa = 5.14AQDD478 pKa = 3.52GDD480 pKa = 4.45NNFKK484 pKa = 11.14FNGTPGIYY492 pKa = 10.04YY493 pKa = 7.49FTVDD497 pKa = 3.94TVNKK501 pKa = 10.23IIDD504 pKa = 3.56LRR506 pKa = 11.84QQ507 pKa = 3.01
MM1 pKa = 7.47KK2 pKa = 10.24KK3 pKa = 9.74ILNICMMVITIIALNSCEE21 pKa = 4.35EE22 pKa = 4.21KK23 pKa = 10.23DD24 pKa = 3.4TSEE27 pKa = 5.74FIAQEE32 pKa = 3.85ATDD35 pKa = 4.16FTFSTSFLEE44 pKa = 5.21NYY46 pKa = 9.92LLNNSLGNNLAEE58 pKa = 4.13RR59 pKa = 11.84FTWRR63 pKa = 11.84DD64 pKa = 3.05ANFGIPTEE72 pKa = 4.16RR73 pKa = 11.84TYY75 pKa = 11.31EE76 pKa = 3.95LQAATMEE83 pKa = 4.68DD84 pKa = 3.62FSDD87 pKa = 3.92YY88 pKa = 11.44TNDD91 pKa = 4.48PDD93 pKa = 4.25DD94 pKa = 4.93SNYY97 pKa = 10.5KK98 pKa = 10.38AYY100 pKa = 11.06NLGTNIGGNEE110 pKa = 3.48IPATVKK116 pKa = 10.5QFLALAKK123 pKa = 10.22IAGLDD128 pKa = 3.72NDD130 pKa = 5.1PEE132 pKa = 4.53TPAPNTGKK140 pKa = 10.75LYY142 pKa = 10.33FRR144 pKa = 11.84VRR146 pKa = 11.84ATIGKK151 pKa = 9.68DD152 pKa = 3.05NLEE155 pKa = 4.24SFSAIQALNIEE166 pKa = 4.26LQEE169 pKa = 4.39GEE171 pKa = 4.55EE172 pKa = 4.25EE173 pKa = 4.38VGGPNCPSWWVVGAGAPDD191 pKa = 4.91AGWNWDD197 pKa = 3.46NPIEE201 pKa = 4.39FLCTDD206 pKa = 3.84QIYY209 pKa = 9.17TSNINLANDD218 pKa = 3.44SFRR221 pKa = 11.84FFTEE225 pKa = 3.29KK226 pKa = 11.07DD227 pKa = 3.12NWDD230 pKa = 3.48SGLNFPYY237 pKa = 10.21FVDD240 pKa = 3.24EE241 pKa = 5.55GYY243 pKa = 10.93SIDD246 pKa = 3.94PNFEE250 pKa = 3.63NAMDD254 pKa = 4.06GDD256 pKa = 4.07EE257 pKa = 4.2NFRR260 pKa = 11.84FIGTPGKK267 pKa = 10.7YY268 pKa = 9.95SLTIDD273 pKa = 4.22SVNKK277 pKa = 9.1TIKK280 pKa = 10.28LGPPAAEE287 pKa = 5.14DD288 pKa = 4.16PNCDD292 pKa = 3.75SVWVVGAGAVDD303 pKa = 5.03AGWNWDD309 pKa = 3.45SPIEE313 pKa = 4.12FFCTDD318 pKa = 2.9NVYY321 pKa = 9.3STNIKK326 pKa = 8.59LTNDD330 pKa = 2.84AFRR333 pKa = 11.84FFLEE337 pKa = 4.0EE338 pKa = 4.39GNWDD342 pKa = 3.38SGQNYY347 pKa = 8.66PFYY350 pKa = 11.24ANDD353 pKa = 4.49GYY355 pKa = 9.72TIDD358 pKa = 5.08SLLEE362 pKa = 4.09DD363 pKa = 4.72AQDD366 pKa = 3.5GDD368 pKa = 4.45NNFKK372 pKa = 11.15FNGTPGTYY380 pKa = 10.55LLTVDD385 pKa = 4.58TVNKK389 pKa = 9.4TITLGIPRR397 pKa = 11.84PEE399 pKa = 4.34EE400 pKa = 4.03PNCDD404 pKa = 3.93SVWVVGAGAVDD415 pKa = 5.03AGWNWDD421 pKa = 3.49SPIEE425 pKa = 4.14FTCTDD430 pKa = 3.32NVYY433 pKa = 10.48SASINLTNDD442 pKa = 2.6AFRR445 pKa = 11.84FFLEE449 pKa = 4.0EE450 pKa = 4.39GNWDD454 pKa = 3.38SGQNYY459 pKa = 8.63PFYY462 pKa = 11.07ADD464 pKa = 4.17DD465 pKa = 5.48GYY467 pKa = 9.64TIDD470 pKa = 4.64TLLEE474 pKa = 4.25DD475 pKa = 5.14AQDD478 pKa = 3.52GDD480 pKa = 4.45NNFKK484 pKa = 11.14FNGTPGIYY492 pKa = 10.04YY493 pKa = 7.49FTVDD497 pKa = 3.94TVNKK501 pKa = 10.23IIDD504 pKa = 3.56LRR506 pKa = 11.84QQ507 pKa = 3.01
Molecular weight: 56.67 kDa
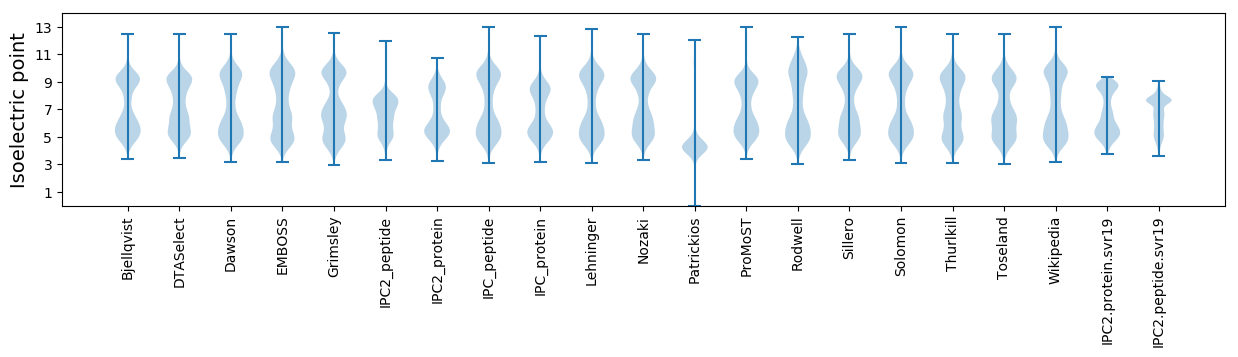
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C4SJ00|A0A5C4SJ00_9FLAO RNA polymerase sigma factor OS=Tamlana fucoidanivorans OX=2583814 GN=FGF67_11925 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1203641 |
24 |
4605 |
361.0 |
40.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.291 ± 0.038 | 0.803 ± 0.014 |
5.655 ± 0.031 | 6.311 ± 0.042 |
5.217 ± 0.035 | 6.469 ± 0.042 |
2.031 ± 0.022 | 7.749 ± 0.037 |
7.791 ± 0.048 | 9.124 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.019 | 6.308 ± 0.043 |
3.498 ± 0.025 | 3.351 ± 0.021 |
3.435 ± 0.028 | 6.42 ± 0.033 |
5.711 ± 0.044 | 6.301 ± 0.033 |
1.188 ± 0.016 | 4.198 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
