
Brevibacterium mcbrellneri ATCC 49030
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Brevibacteriaceae; Brevibacterium; Brevibacterium mcbrellneri
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
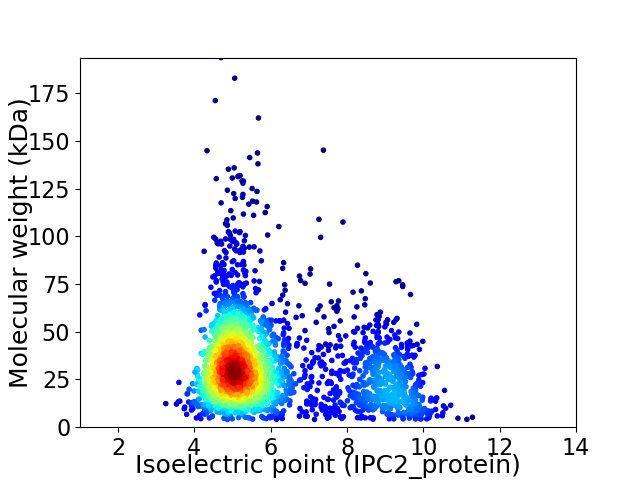
Virtual 2D-PAGE plot for 2432 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4YQJ4|D4YQJ4_9MICO Uncharacterized protein OS=Brevibacterium mcbrellneri ATCC 49030 OX=585530 GN=HMPREF0183_2204 PE=4 SV=1
MM1 pKa = 7.6ARR3 pKa = 11.84RR4 pKa = 11.84ATLPILIFLLMTSGCSEE21 pKa = 3.77NSGVDD26 pKa = 3.35VHH28 pKa = 7.54SDD30 pKa = 3.2SPEE33 pKa = 3.72ISVGATSAHH42 pKa = 6.14SLPPITGVFDD52 pKa = 3.76YY53 pKa = 10.93QLGGAYY59 pKa = 10.3SLPTPIDD66 pKa = 3.47VVVRR70 pKa = 11.84DD71 pKa = 4.47ASAEE75 pKa = 4.07PLSDD79 pKa = 3.77AYY81 pKa = 10.28SVCYY85 pKa = 10.85VNGFQTQPDD94 pKa = 3.98DD95 pKa = 3.24AAMWKK100 pKa = 9.36EE101 pKa = 3.82HH102 pKa = 7.07ADD104 pKa = 3.53LLLHH108 pKa = 6.42NKK110 pKa = 10.06HH111 pKa = 6.34GDD113 pKa = 3.57LVTDD117 pKa = 4.66PDD119 pKa = 3.71WDD121 pKa = 3.78DD122 pKa = 4.95EE123 pKa = 4.83YY124 pKa = 10.99ILDD127 pKa = 5.03PSTPDD132 pKa = 3.03QRR134 pKa = 11.84ADD136 pKa = 2.94ILKK139 pKa = 9.73IISPIITGCANAGYY153 pKa = 10.18DD154 pKa = 4.07AVEE157 pKa = 4.26IDD159 pKa = 4.15NLDD162 pKa = 3.44SFDD165 pKa = 4.34RR166 pKa = 11.84FAQIEE171 pKa = 4.19QNAAMTLASAYY182 pKa = 10.73VEE184 pKa = 4.25IAHH187 pKa = 7.09DD188 pKa = 4.31AGLAIAQKK196 pKa = 10.2NAAEE200 pKa = 4.52IARR203 pKa = 11.84SAHH206 pKa = 6.4EE207 pKa = 4.44DD208 pKa = 3.27LGFDD212 pKa = 3.7FAVTEE217 pKa = 4.23EE218 pKa = 4.26CGSYY222 pKa = 10.55DD223 pKa = 3.27EE224 pKa = 5.19CDD226 pKa = 3.53AYY228 pKa = 10.12TSVYY232 pKa = 9.39GPHH235 pKa = 6.56VLEE238 pKa = 4.77IEE240 pKa = 4.47YY241 pKa = 10.22PDD243 pKa = 3.8SLEE246 pKa = 3.99EE247 pKa = 5.03AGMTFADD254 pKa = 4.12VCALPDD260 pKa = 3.85RR261 pKa = 11.84APLTILRR268 pKa = 11.84DD269 pKa = 3.41RR270 pKa = 11.84ALVARR275 pKa = 11.84GAKK278 pKa = 9.9GYY280 pKa = 10.35RR281 pKa = 11.84LEE283 pKa = 4.28SCC285 pKa = 4.39
MM1 pKa = 7.6ARR3 pKa = 11.84RR4 pKa = 11.84ATLPILIFLLMTSGCSEE21 pKa = 3.77NSGVDD26 pKa = 3.35VHH28 pKa = 7.54SDD30 pKa = 3.2SPEE33 pKa = 3.72ISVGATSAHH42 pKa = 6.14SLPPITGVFDD52 pKa = 3.76YY53 pKa = 10.93QLGGAYY59 pKa = 10.3SLPTPIDD66 pKa = 3.47VVVRR70 pKa = 11.84DD71 pKa = 4.47ASAEE75 pKa = 4.07PLSDD79 pKa = 3.77AYY81 pKa = 10.28SVCYY85 pKa = 10.85VNGFQTQPDD94 pKa = 3.98DD95 pKa = 3.24AAMWKK100 pKa = 9.36EE101 pKa = 3.82HH102 pKa = 7.07ADD104 pKa = 3.53LLLHH108 pKa = 6.42NKK110 pKa = 10.06HH111 pKa = 6.34GDD113 pKa = 3.57LVTDD117 pKa = 4.66PDD119 pKa = 3.71WDD121 pKa = 3.78DD122 pKa = 4.95EE123 pKa = 4.83YY124 pKa = 10.99ILDD127 pKa = 5.03PSTPDD132 pKa = 3.03QRR134 pKa = 11.84ADD136 pKa = 2.94ILKK139 pKa = 9.73IISPIITGCANAGYY153 pKa = 10.18DD154 pKa = 4.07AVEE157 pKa = 4.26IDD159 pKa = 4.15NLDD162 pKa = 3.44SFDD165 pKa = 4.34RR166 pKa = 11.84FAQIEE171 pKa = 4.19QNAAMTLASAYY182 pKa = 10.73VEE184 pKa = 4.25IAHH187 pKa = 7.09DD188 pKa = 4.31AGLAIAQKK196 pKa = 10.2NAAEE200 pKa = 4.52IARR203 pKa = 11.84SAHH206 pKa = 6.4EE207 pKa = 4.44DD208 pKa = 3.27LGFDD212 pKa = 3.7FAVTEE217 pKa = 4.23EE218 pKa = 4.26CGSYY222 pKa = 10.55DD223 pKa = 3.27EE224 pKa = 5.19CDD226 pKa = 3.53AYY228 pKa = 10.12TSVYY232 pKa = 9.39GPHH235 pKa = 6.56VLEE238 pKa = 4.77IEE240 pKa = 4.47YY241 pKa = 10.22PDD243 pKa = 3.8SLEE246 pKa = 3.99EE247 pKa = 5.03AGMTFADD254 pKa = 4.12VCALPDD260 pKa = 3.85RR261 pKa = 11.84APLTILRR268 pKa = 11.84DD269 pKa = 3.41RR270 pKa = 11.84ALVARR275 pKa = 11.84GAKK278 pKa = 9.9GYY280 pKa = 10.35RR281 pKa = 11.84LEE283 pKa = 4.28SCC285 pKa = 4.39
Molecular weight: 30.78 kDa
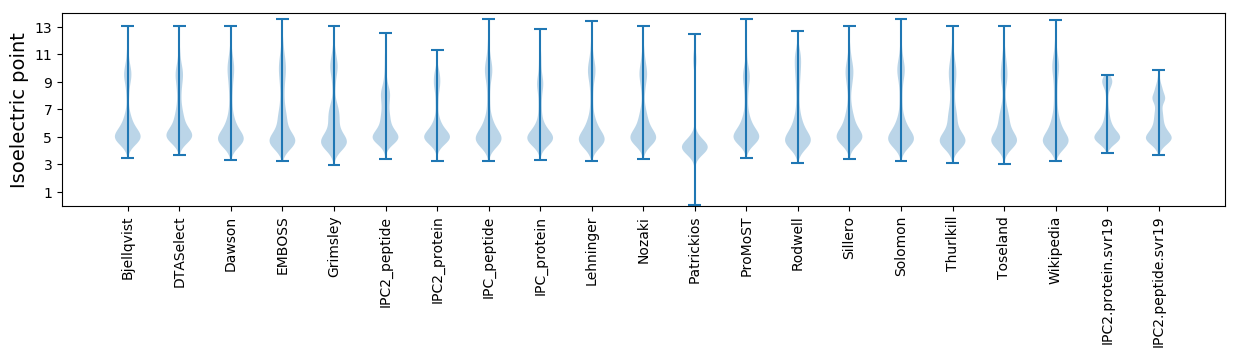
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4YKW3|D4YKW3_9MICO tRNA adenylyltransferase OS=Brevibacterium mcbrellneri ATCC 49030 OX=585530 GN=HMPREF0183_0573 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84KK12 pKa = 9.72RR13 pKa = 11.84AKK15 pKa = 8.66THH17 pKa = 5.25GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84KK12 pKa = 9.72RR13 pKa = 11.84AKK15 pKa = 8.66THH17 pKa = 5.25GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
779593 |
32 |
1818 |
320.6 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.036 ± 0.067 | 0.701 ± 0.013 |
5.98 ± 0.04 | 6.195 ± 0.052 |
3.365 ± 0.032 | 8.32 ± 0.045 |
2.292 ± 0.023 | 4.782 ± 0.035 |
3.544 ± 0.048 | 9.302 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.171 ± 0.021 | 2.86 ± 0.028 |
5.079 ± 0.037 | 3.376 ± 0.03 |
6.124 ± 0.041 | 6.121 ± 0.033 |
6.491 ± 0.039 | 8.799 ± 0.051 |
1.314 ± 0.021 | 2.147 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
