
Xylaria hypoxylon
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariaceae; Xylaria
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
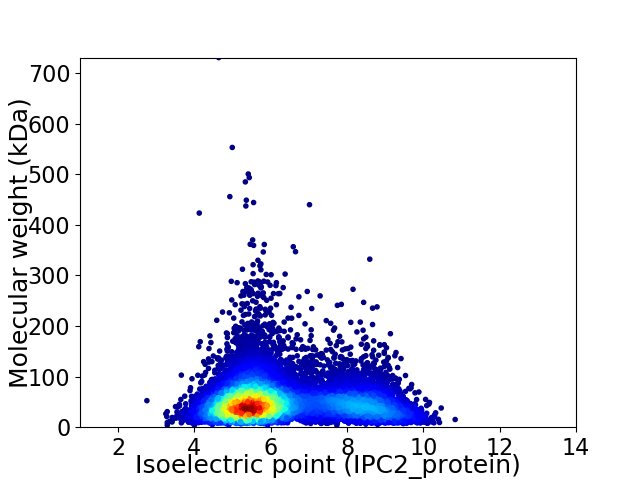
Virtual 2D-PAGE plot for 11037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0Z8W8|A0A4Z0Z8W8_9PEZI tRNA wybutosine-synthesizing protein 4 OS=Xylaria hypoxylon OX=37992 GN=E0Z10_g2906 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 10.0PNEE6 pKa = 4.42AIAVVVTVLILGGFIAFYY24 pKa = 10.95VFTYY28 pKa = 9.61CFRR31 pKa = 11.84HH32 pKa = 6.65SGDD35 pKa = 5.32DD36 pKa = 5.33DD37 pKa = 6.68DD38 pKa = 7.69DD39 pKa = 7.31DD40 pKa = 7.65DD41 pKa = 6.89DD42 pKa = 6.71DD43 pKa = 5.51EE44 pKa = 4.98YY45 pKa = 11.88NNDD48 pKa = 3.32YY49 pKa = 11.08GAVGSSAFLSTSTSTSAGEE68 pKa = 4.11PGDD71 pKa = 4.58GILRR75 pKa = 11.84PPPVVFAQSGRR86 pKa = 11.84SQYY89 pKa = 10.69PPANTSFPIGTDD101 pKa = 3.35TVPDD105 pKa = 4.61LPYY108 pKa = 10.45QDD110 pKa = 4.75NGTAAGGAGSHH121 pKa = 6.31GNGVGIAEE129 pKa = 4.65GTSGTPPPPAVDD141 pKa = 3.52VEE143 pKa = 4.43HH144 pKa = 7.34PDD146 pKa = 3.94DD147 pKa = 4.7RR148 pKa = 11.84GG149 pKa = 3.18
MM1 pKa = 7.55TKK3 pKa = 10.0PNEE6 pKa = 4.42AIAVVVTVLILGGFIAFYY24 pKa = 10.95VFTYY28 pKa = 9.61CFRR31 pKa = 11.84HH32 pKa = 6.65SGDD35 pKa = 5.32DD36 pKa = 5.33DD37 pKa = 6.68DD38 pKa = 7.69DD39 pKa = 7.31DD40 pKa = 7.65DD41 pKa = 6.89DD42 pKa = 6.71DD43 pKa = 5.51EE44 pKa = 4.98YY45 pKa = 11.88NNDD48 pKa = 3.32YY49 pKa = 11.08GAVGSSAFLSTSTSTSAGEE68 pKa = 4.11PGDD71 pKa = 4.58GILRR75 pKa = 11.84PPPVVFAQSGRR86 pKa = 11.84SQYY89 pKa = 10.69PPANTSFPIGTDD101 pKa = 3.35TVPDD105 pKa = 4.61LPYY108 pKa = 10.45QDD110 pKa = 4.75NGTAAGGAGSHH121 pKa = 6.31GNGVGIAEE129 pKa = 4.65GTSGTPPPPAVDD141 pKa = 3.52VEE143 pKa = 4.43HH144 pKa = 7.34PDD146 pKa = 3.94DD147 pKa = 4.7RR148 pKa = 11.84GG149 pKa = 3.18
Molecular weight: 15.31 kDa
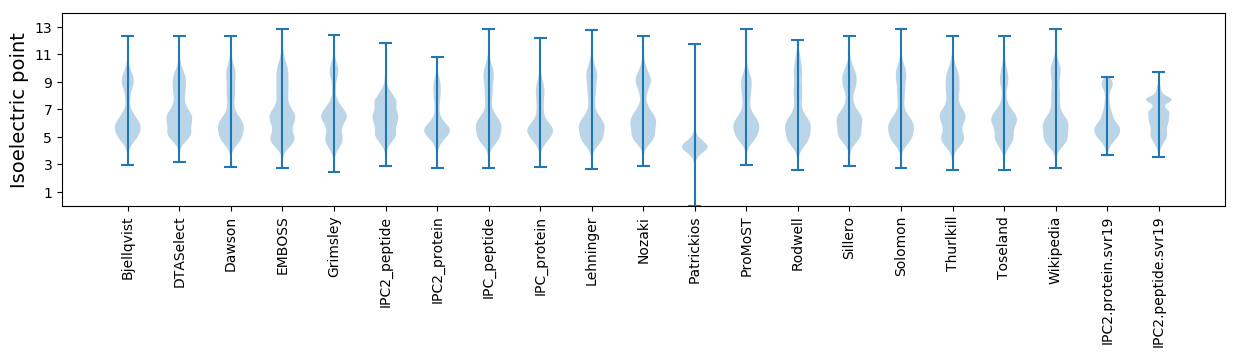
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0YLJ0|A0A4Z0YLJ0_9PEZI GATA-type domain-containing protein OS=Xylaria hypoxylon OX=37992 GN=E0Z10_g8450 PE=4 SV=1
MM1 pKa = 7.05MSSFRR6 pKa = 11.84GLCLLRR12 pKa = 11.84ATQTRR17 pKa = 11.84PSHH20 pKa = 6.18IPSSLHH26 pKa = 6.0LRR28 pKa = 11.84PSSITSRR35 pKa = 11.84ISPSPLHH42 pKa = 6.95HH43 pKa = 6.43IQQHH47 pKa = 4.74GRR49 pKa = 11.84RR50 pKa = 11.84SFHH53 pKa = 5.54WQSAASTTIEE63 pKa = 4.12GAQNLIVDD71 pKa = 4.35LHH73 pKa = 6.12TVTSLPWFLTIPLVAFTVGAVFRR96 pKa = 11.84LPFSIYY102 pKa = 7.03TQRR105 pKa = 11.84ILQRR109 pKa = 11.84RR110 pKa = 11.84TNFGPLLQAWNARR123 pKa = 11.84IQQDD127 pKa = 3.76VQQEE131 pKa = 4.69GISASSRR138 pKa = 11.84MSEE141 pKa = 3.8VKK143 pKa = 10.35ARR145 pKa = 11.84QDD147 pKa = 3.03KK148 pKa = 10.11ALKK151 pKa = 10.15RR152 pKa = 11.84IYY154 pKa = 10.18RR155 pKa = 11.84KK156 pKa = 10.25LGLQEE161 pKa = 3.1WRR163 pKa = 11.84MWGSILSFPIWLVAIDD179 pKa = 3.89AVRR182 pKa = 11.84RR183 pKa = 11.84LCGGPRR189 pKa = 11.84GLIGSLFIGPGSSSEE204 pKa = 4.29TTVAAHH210 pKa = 6.15VEE212 pKa = 4.38AASSLPPGSVADD224 pKa = 4.58PSTLDD229 pKa = 3.37PVAISSAVEE238 pKa = 3.93TAHH241 pKa = 6.51MATVDD246 pKa = 3.61PSLTLEE252 pKa = 4.24GCLWFTDD259 pKa = 3.98LTASDD264 pKa = 5.15PYY266 pKa = 11.41HH267 pKa = 6.45MLPIALSVTLVLNMLPKK284 pKa = 10.4SGEE287 pKa = 4.0KK288 pKa = 10.4FSDD291 pKa = 3.12RR292 pKa = 11.84VRR294 pKa = 11.84IALGRR299 pKa = 11.84RR300 pKa = 11.84PKK302 pKa = 10.27SARR305 pKa = 11.84AQTFAGDD312 pKa = 3.53EE313 pKa = 4.07KK314 pKa = 11.09VGFRR318 pKa = 11.84EE319 pKa = 3.77RR320 pKa = 11.84LFATFYY326 pKa = 10.76LCMVGVATLVGPLTLDD342 pKa = 3.76LPAALHH348 pKa = 6.28LYY350 pKa = 8.69WLASSMSNVLFMKK363 pKa = 10.21GLRR366 pKa = 11.84HH367 pKa = 6.05LMPVEE372 pKa = 3.81GRR374 pKa = 11.84LLKK377 pKa = 10.51RR378 pKa = 11.84CTGAEE383 pKa = 3.99FPVIRR388 pKa = 11.84PQRR391 pKa = 11.84QQKK394 pKa = 8.86NN395 pKa = 3.3
MM1 pKa = 7.05MSSFRR6 pKa = 11.84GLCLLRR12 pKa = 11.84ATQTRR17 pKa = 11.84PSHH20 pKa = 6.18IPSSLHH26 pKa = 6.0LRR28 pKa = 11.84PSSITSRR35 pKa = 11.84ISPSPLHH42 pKa = 6.95HH43 pKa = 6.43IQQHH47 pKa = 4.74GRR49 pKa = 11.84RR50 pKa = 11.84SFHH53 pKa = 5.54WQSAASTTIEE63 pKa = 4.12GAQNLIVDD71 pKa = 4.35LHH73 pKa = 6.12TVTSLPWFLTIPLVAFTVGAVFRR96 pKa = 11.84LPFSIYY102 pKa = 7.03TQRR105 pKa = 11.84ILQRR109 pKa = 11.84RR110 pKa = 11.84TNFGPLLQAWNARR123 pKa = 11.84IQQDD127 pKa = 3.76VQQEE131 pKa = 4.69GISASSRR138 pKa = 11.84MSEE141 pKa = 3.8VKK143 pKa = 10.35ARR145 pKa = 11.84QDD147 pKa = 3.03KK148 pKa = 10.11ALKK151 pKa = 10.15RR152 pKa = 11.84IYY154 pKa = 10.18RR155 pKa = 11.84KK156 pKa = 10.25LGLQEE161 pKa = 3.1WRR163 pKa = 11.84MWGSILSFPIWLVAIDD179 pKa = 3.89AVRR182 pKa = 11.84RR183 pKa = 11.84LCGGPRR189 pKa = 11.84GLIGSLFIGPGSSSEE204 pKa = 4.29TTVAAHH210 pKa = 6.15VEE212 pKa = 4.38AASSLPPGSVADD224 pKa = 4.58PSTLDD229 pKa = 3.37PVAISSAVEE238 pKa = 3.93TAHH241 pKa = 6.51MATVDD246 pKa = 3.61PSLTLEE252 pKa = 4.24GCLWFTDD259 pKa = 3.98LTASDD264 pKa = 5.15PYY266 pKa = 11.41HH267 pKa = 6.45MLPIALSVTLVLNMLPKK284 pKa = 10.4SGEE287 pKa = 4.0KK288 pKa = 10.4FSDD291 pKa = 3.12RR292 pKa = 11.84VRR294 pKa = 11.84IALGRR299 pKa = 11.84RR300 pKa = 11.84PKK302 pKa = 10.27SARR305 pKa = 11.84AQTFAGDD312 pKa = 3.53EE313 pKa = 4.07KK314 pKa = 11.09VGFRR318 pKa = 11.84EE319 pKa = 3.77RR320 pKa = 11.84LFATFYY326 pKa = 10.76LCMVGVATLVGPLTLDD342 pKa = 3.76LPAALHH348 pKa = 6.28LYY350 pKa = 8.69WLASSMSNVLFMKK363 pKa = 10.21GLRR366 pKa = 11.84HH367 pKa = 6.05LMPVEE372 pKa = 3.81GRR374 pKa = 11.84LLKK377 pKa = 10.51RR378 pKa = 11.84CTGAEE383 pKa = 3.99FPVIRR388 pKa = 11.84PQRR391 pKa = 11.84QQKK394 pKa = 8.86NN395 pKa = 3.3
Molecular weight: 43.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5574772 |
35 |
6685 |
505.1 |
55.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.488 ± 0.016 | 1.137 ± 0.008 |
5.876 ± 0.016 | 6.112 ± 0.022 |
3.725 ± 0.013 | 6.855 ± 0.02 |
2.398 ± 0.009 | 5.137 ± 0.014 |
4.767 ± 0.02 | 8.912 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.009 | 3.843 ± 0.012 |
5.932 ± 0.023 | 3.957 ± 0.014 |
5.998 ± 0.018 | 8.24 ± 0.024 |
6.141 ± 0.014 | 6.142 ± 0.014 |
1.465 ± 0.009 | 2.809 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
