
Tortoise microvirus 67
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
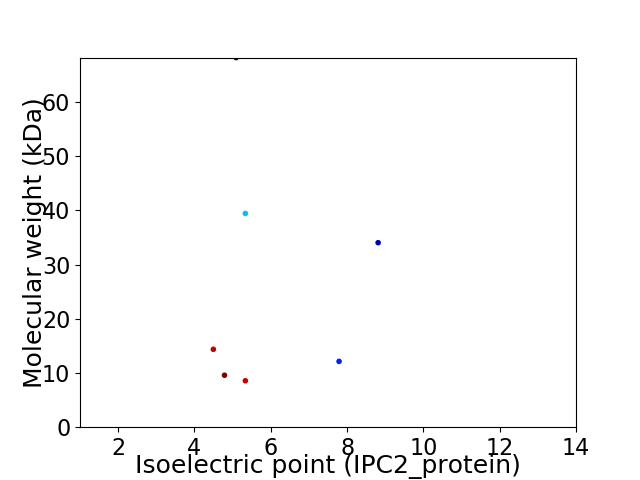
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
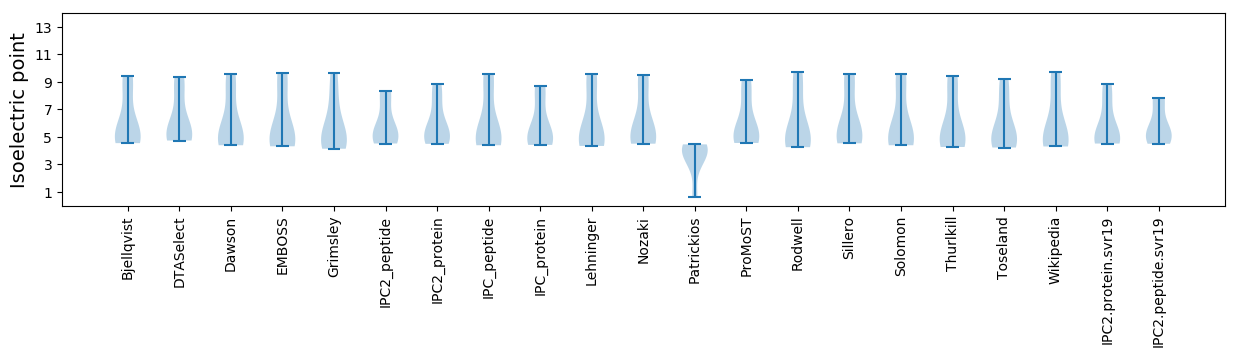
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6L0|A0A4P8W6L0_9VIRU Replication initiation protein OS=Tortoise microvirus 67 OX=2583173 PE=4 SV=1
MM1 pKa = 7.37LRR3 pKa = 11.84YY4 pKa = 10.12DD5 pKa = 5.01GISSLGGCNLVEE17 pKa = 4.14YY18 pKa = 10.31DD19 pKa = 4.72PPYY22 pKa = 10.93LDD24 pKa = 4.63ADD26 pKa = 4.11FGEE29 pKa = 4.79SVTDD33 pKa = 3.65DD34 pKa = 3.29SYY36 pKa = 12.02YY37 pKa = 10.26IPTPDD42 pKa = 3.31NVGLKK47 pKa = 10.11SIPLTSSEE55 pKa = 4.15ILSNFDD61 pKa = 3.39FSDD64 pKa = 3.63GRR66 pKa = 11.84DD67 pKa = 3.18TGAPVPKK74 pKa = 9.73FRR76 pKa = 11.84KK77 pKa = 9.35PGADD81 pKa = 3.05LGEE84 pKa = 4.41VSQHH88 pKa = 4.87LRR90 pKa = 11.84EE91 pKa = 4.14VQGEE95 pKa = 4.27IKK97 pKa = 10.7SEE99 pKa = 4.2VARR102 pKa = 11.84KK103 pKa = 8.95AAEE106 pKa = 3.71KK107 pKa = 10.78ALADD111 pKa = 3.9KK112 pKa = 11.28YY113 pKa = 11.12KK114 pKa = 10.16VTDD117 pKa = 3.94SPDD120 pKa = 3.49TPHH123 pKa = 6.96APSVAASPNSVV134 pKa = 2.73
MM1 pKa = 7.37LRR3 pKa = 11.84YY4 pKa = 10.12DD5 pKa = 5.01GISSLGGCNLVEE17 pKa = 4.14YY18 pKa = 10.31DD19 pKa = 4.72PPYY22 pKa = 10.93LDD24 pKa = 4.63ADD26 pKa = 4.11FGEE29 pKa = 4.79SVTDD33 pKa = 3.65DD34 pKa = 3.29SYY36 pKa = 12.02YY37 pKa = 10.26IPTPDD42 pKa = 3.31NVGLKK47 pKa = 10.11SIPLTSSEE55 pKa = 4.15ILSNFDD61 pKa = 3.39FSDD64 pKa = 3.63GRR66 pKa = 11.84DD67 pKa = 3.18TGAPVPKK74 pKa = 9.73FRR76 pKa = 11.84KK77 pKa = 9.35PGADD81 pKa = 3.05LGEE84 pKa = 4.41VSQHH88 pKa = 4.87LRR90 pKa = 11.84EE91 pKa = 4.14VQGEE95 pKa = 4.27IKK97 pKa = 10.7SEE99 pKa = 4.2VARR102 pKa = 11.84KK103 pKa = 8.95AAEE106 pKa = 3.71KK107 pKa = 10.78ALADD111 pKa = 3.9KK112 pKa = 11.28YY113 pKa = 11.12KK114 pKa = 10.16VTDD117 pKa = 3.94SPDD120 pKa = 3.49TPHH123 pKa = 6.96APSVAASPNSVV134 pKa = 2.73
Molecular weight: 14.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6Y3|A0A4P8W6Y3_9VIRU Uncharacterized protein OS=Tortoise microvirus 67 OX=2583173 PE=4 SV=1
MM1 pKa = 7.45SCLFPVTVYY10 pKa = 10.92PNGKK14 pKa = 9.05KK15 pKa = 10.23APPVEE20 pKa = 4.45APCRR24 pKa = 11.84HH25 pKa = 6.31CANCRR30 pKa = 11.84MAKK33 pKa = 10.1QITLNFAIGHH43 pKa = 7.04ALQDD47 pKa = 3.86CYY49 pKa = 11.72SRR51 pKa = 11.84GGSASFVTMTYY62 pKa = 11.08ADD64 pKa = 3.75QFLPDD69 pKa = 3.77NGSLRR74 pKa = 11.84SKK76 pKa = 11.28DD77 pKa = 3.16FTNWLKK83 pKa = 10.77GARR86 pKa = 11.84TKK88 pKa = 10.88LFRR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 9.11IDD95 pKa = 3.54LPFKK99 pKa = 10.0YY100 pKa = 9.74LAAGEE105 pKa = 4.21YY106 pKa = 10.0GGKK109 pKa = 9.95LGRR112 pKa = 11.84PHH114 pKa = 5.56YY115 pKa = 10.23HH116 pKa = 7.2CILIGIPRR124 pKa = 11.84PVAEE128 pKa = 4.51GVFRR132 pKa = 11.84PLWRR136 pKa = 11.84HH137 pKa = 5.05GHH139 pKa = 5.69CQTEE143 pKa = 4.35VLGSGGVRR151 pKa = 11.84YY152 pKa = 10.08VSEE155 pKa = 4.62YY156 pKa = 9.89ISKK159 pKa = 10.71QIGGRR164 pKa = 11.84LAEE167 pKa = 3.95QMYY170 pKa = 9.79DD171 pKa = 3.11AKK173 pKa = 10.98GLEE176 pKa = 4.14RR177 pKa = 11.84PFMRR181 pKa = 11.84RR182 pKa = 11.84SVNFCDD188 pKa = 4.78DD189 pKa = 4.64WILKK193 pKa = 10.21NYY195 pKa = 10.07ADD197 pKa = 4.2LVSNGWLFKK206 pKa = 10.91SRR208 pKa = 11.84GKK210 pKa = 9.21MRR212 pKa = 11.84PVPSYY217 pKa = 10.53YY218 pKa = 7.95VHH220 pKa = 7.37KK221 pKa = 10.93YY222 pKa = 10.2FGDD225 pKa = 3.55VDD227 pKa = 3.96LHH229 pKa = 6.73FLAQEE234 pKa = 3.93RR235 pKa = 11.84NLAAKK240 pKa = 8.94RR241 pKa = 11.84AKK243 pKa = 9.72MSLTDD248 pKa = 3.46YY249 pKa = 11.25EE250 pKa = 4.49FMRR253 pKa = 11.84NYY255 pKa = 9.2TLEE258 pKa = 4.34GMSHH262 pKa = 6.4RR263 pKa = 11.84ASRR266 pKa = 11.84AKK268 pKa = 10.46GVASDD273 pKa = 3.58SSSYY277 pKa = 11.67DD278 pKa = 3.02LMPFHH283 pKa = 7.26SSCDD287 pKa = 3.19IASLVKK293 pKa = 10.59DD294 pKa = 4.02ALDD297 pKa = 3.59PVPFF301 pKa = 4.8
MM1 pKa = 7.45SCLFPVTVYY10 pKa = 10.92PNGKK14 pKa = 9.05KK15 pKa = 10.23APPVEE20 pKa = 4.45APCRR24 pKa = 11.84HH25 pKa = 6.31CANCRR30 pKa = 11.84MAKK33 pKa = 10.1QITLNFAIGHH43 pKa = 7.04ALQDD47 pKa = 3.86CYY49 pKa = 11.72SRR51 pKa = 11.84GGSASFVTMTYY62 pKa = 11.08ADD64 pKa = 3.75QFLPDD69 pKa = 3.77NGSLRR74 pKa = 11.84SKK76 pKa = 11.28DD77 pKa = 3.16FTNWLKK83 pKa = 10.77GARR86 pKa = 11.84TKK88 pKa = 10.88LFRR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 9.11IDD95 pKa = 3.54LPFKK99 pKa = 10.0YY100 pKa = 9.74LAAGEE105 pKa = 4.21YY106 pKa = 10.0GGKK109 pKa = 9.95LGRR112 pKa = 11.84PHH114 pKa = 5.56YY115 pKa = 10.23HH116 pKa = 7.2CILIGIPRR124 pKa = 11.84PVAEE128 pKa = 4.51GVFRR132 pKa = 11.84PLWRR136 pKa = 11.84HH137 pKa = 5.05GHH139 pKa = 5.69CQTEE143 pKa = 4.35VLGSGGVRR151 pKa = 11.84YY152 pKa = 10.08VSEE155 pKa = 4.62YY156 pKa = 9.89ISKK159 pKa = 10.71QIGGRR164 pKa = 11.84LAEE167 pKa = 3.95QMYY170 pKa = 9.79DD171 pKa = 3.11AKK173 pKa = 10.98GLEE176 pKa = 4.14RR177 pKa = 11.84PFMRR181 pKa = 11.84RR182 pKa = 11.84SVNFCDD188 pKa = 4.78DD189 pKa = 4.64WILKK193 pKa = 10.21NYY195 pKa = 10.07ADD197 pKa = 4.2LVSNGWLFKK206 pKa = 10.91SRR208 pKa = 11.84GKK210 pKa = 9.21MRR212 pKa = 11.84PVPSYY217 pKa = 10.53YY218 pKa = 7.95VHH220 pKa = 7.37KK221 pKa = 10.93YY222 pKa = 10.2FGDD225 pKa = 3.55VDD227 pKa = 3.96LHH229 pKa = 6.73FLAQEE234 pKa = 3.93RR235 pKa = 11.84NLAAKK240 pKa = 8.94RR241 pKa = 11.84AKK243 pKa = 9.72MSLTDD248 pKa = 3.46YY249 pKa = 11.25EE250 pKa = 4.49FMRR253 pKa = 11.84NYY255 pKa = 9.2TLEE258 pKa = 4.34GMSHH262 pKa = 6.4RR263 pKa = 11.84ASRR266 pKa = 11.84AKK268 pKa = 10.46GVASDD273 pKa = 3.58SSSYY277 pKa = 11.67DD278 pKa = 3.02LMPFHH283 pKa = 7.26SSCDD287 pKa = 3.19IASLVKK293 pKa = 10.59DD294 pKa = 4.02ALDD297 pKa = 3.59PVPFF301 pKa = 4.8
Molecular weight: 34.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1670 |
77 |
614 |
238.6 |
26.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.323 ± 0.682 | 1.557 ± 0.615 |
5.868 ± 0.453 | 4.97 ± 0.66 |
5.928 ± 0.786 | 6.886 ± 0.485 |
1.617 ± 0.376 | 4.85 ± 0.493 |
3.832 ± 0.833 | 8.263 ± 0.585 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.335 ± 0.273 | 4.311 ± 0.462 |
5.09 ± 0.797 | 4.012 ± 1.493 |
6.048 ± 0.631 | 9.281 ± 0.689 |
5.389 ± 0.974 | 5.689 ± 0.632 |
0.719 ± 0.178 | 5.03 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
