
Candidatus Magnetoovum chiemensis
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Candidatus Magnetoovum
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
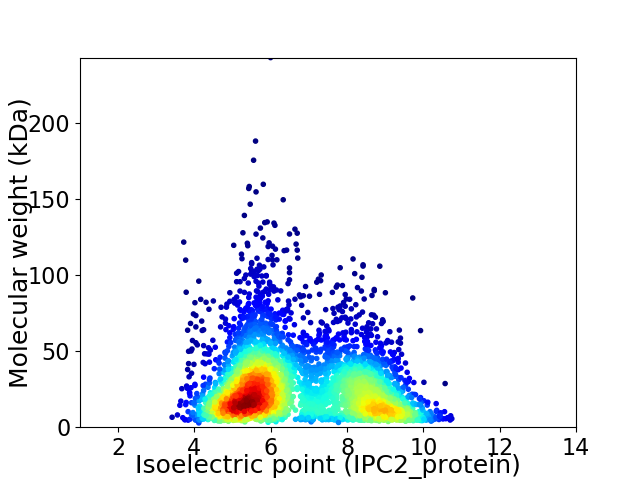
Virtual 2D-PAGE plot for 4107 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F2IY09|A0A0F2IY09_9BACT Nitrogenase molybdenum-iron protein beta chain OS=Candidatus Magnetoovum chiemensis OX=1609970 GN=MCHI_003117 PE=3 SV=1
MM1 pKa = 7.49TIAGEE6 pKa = 4.49TFTITQDD13 pKa = 2.84PCTYY17 pKa = 10.34SIQPEE22 pKa = 4.64TKK24 pKa = 9.57SFTYY28 pKa = 9.36TGGVGFVTINTSEE41 pKa = 4.15GCTWEE46 pKa = 4.66AVSNDD51 pKa = 2.93LWLSLMSGTSGAGSGQAIFAVEE73 pKa = 4.18TNDD76 pKa = 3.59ASASRR81 pKa = 11.84TGTLTIAGEE90 pKa = 4.56TFTVTQSATDD100 pKa = 3.72CEE102 pKa = 4.6YY103 pKa = 11.08SISPSKK109 pKa = 10.81ASYY112 pKa = 8.84STDD115 pKa = 2.8GGAGTIEE122 pKa = 4.25VSSSTGCEE130 pKa = 3.89WTPEE134 pKa = 4.04ASDD137 pKa = 3.53DD138 pKa = 4.24WISITSGSSGNGIGTITYY156 pKa = 7.51TVNSNSGFQRR166 pKa = 11.84GGKK169 pKa = 8.83IEE171 pKa = 4.03IMGEE175 pKa = 3.87EE176 pKa = 4.02VDD178 pKa = 4.1IVQAGDD184 pKa = 3.38PCVYY188 pKa = 11.16DD189 pKa = 4.3LMPKK193 pKa = 9.66VGVYY197 pKa = 8.98TSQSGSGSVDD207 pKa = 3.43VITSLDD213 pKa = 3.35TCAWTAVSDD222 pKa = 4.06NSWITIDD229 pKa = 3.89AGSNGTGNAAVSYY242 pKa = 10.31SVSEE246 pKa = 4.34NPNTSPRR253 pKa = 11.84IGILEE258 pKa = 3.66IAGRR262 pKa = 11.84KK263 pKa = 6.73FTVTQTGADD272 pKa = 3.66ATCSVSILPQQTTISSEE289 pKa = 4.06STEE292 pKa = 4.0GNIIVLTSPADD303 pKa = 4.04CAWSTTTVASSWIAISDD320 pKa = 4.01PEE322 pKa = 4.52SGSGTGDD329 pKa = 3.3SVITYY334 pKa = 9.56KK335 pKa = 11.02AEE337 pKa = 4.19ANADD341 pKa = 3.31TSDD344 pKa = 3.39RR345 pKa = 11.84FGVVMVNDD353 pKa = 3.76QPFMLTQKK361 pKa = 10.3GVCTYY366 pKa = 10.66DD367 pKa = 3.68LSPVVIDD374 pKa = 3.91IDD376 pKa = 3.91AAGSASEE383 pKa = 4.39TVAVTPSDD391 pKa = 5.56SICTWNAISDD401 pKa = 4.14EE402 pKa = 4.1DD403 pKa = 4.32WITVVSSSSSGTGTGTVEE421 pKa = 3.89FAVAANSTGAMRR433 pKa = 11.84TGKK436 pKa = 10.12VVIGGQNLLIVQLSEE451 pKa = 3.73AGKK454 pKa = 9.56PFDD457 pKa = 6.42DD458 pKa = 5.14IDD460 pKa = 4.97SEE462 pKa = 5.65DD463 pKa = 3.82DD464 pKa = 3.48TFQNAINAMKK474 pKa = 10.06AYY476 pKa = 10.14SITEE480 pKa = 4.03GCDD483 pKa = 3.15AQGNYY488 pKa = 9.83CPEE491 pKa = 4.59GYY493 pKa = 10.67VNRR496 pKa = 11.84AQMAVFVLKK505 pKa = 10.64AAGQAPSDD513 pKa = 3.77SDD515 pKa = 3.8TCTGAMFNDD524 pKa = 3.75VTTEE528 pKa = 3.77TLGDD532 pKa = 3.94YY533 pKa = 9.89FCKK536 pKa = 10.48AIEE539 pKa = 4.5KK540 pKa = 9.76FASLNITAGCGDD552 pKa = 4.37GNFCPDD558 pKa = 3.36GYY560 pKa = 8.94VTRR563 pKa = 11.84AQMAVFMLKK572 pKa = 10.72GLGQGPNSSDD582 pKa = 2.99KK583 pKa = 11.48CKK585 pKa = 10.08GTEE588 pKa = 4.24LNDD591 pKa = 2.96ITTDD595 pKa = 3.12KK596 pKa = 11.02VGEE599 pKa = 4.25YY600 pKa = 9.79FCKK603 pKa = 10.49AIEE606 pKa = 4.06KK607 pKa = 10.34FRR609 pKa = 11.84DD610 pKa = 3.33KK611 pKa = 11.19NITTGCSVDD620 pKa = 5.26DD621 pKa = 4.28PLTTEE626 pKa = 4.95NEE628 pKa = 3.78AAYY631 pKa = 10.74CPDD634 pKa = 4.06DD635 pKa = 3.75NVKK638 pKa = 10.11RR639 pKa = 11.84SQMALFLTKK648 pKa = 10.72GFLSDD653 pKa = 3.49KK654 pKa = 11.03
MM1 pKa = 7.49TIAGEE6 pKa = 4.49TFTITQDD13 pKa = 2.84PCTYY17 pKa = 10.34SIQPEE22 pKa = 4.64TKK24 pKa = 9.57SFTYY28 pKa = 9.36TGGVGFVTINTSEE41 pKa = 4.15GCTWEE46 pKa = 4.66AVSNDD51 pKa = 2.93LWLSLMSGTSGAGSGQAIFAVEE73 pKa = 4.18TNDD76 pKa = 3.59ASASRR81 pKa = 11.84TGTLTIAGEE90 pKa = 4.56TFTVTQSATDD100 pKa = 3.72CEE102 pKa = 4.6YY103 pKa = 11.08SISPSKK109 pKa = 10.81ASYY112 pKa = 8.84STDD115 pKa = 2.8GGAGTIEE122 pKa = 4.25VSSSTGCEE130 pKa = 3.89WTPEE134 pKa = 4.04ASDD137 pKa = 3.53DD138 pKa = 4.24WISITSGSSGNGIGTITYY156 pKa = 7.51TVNSNSGFQRR166 pKa = 11.84GGKK169 pKa = 8.83IEE171 pKa = 4.03IMGEE175 pKa = 3.87EE176 pKa = 4.02VDD178 pKa = 4.1IVQAGDD184 pKa = 3.38PCVYY188 pKa = 11.16DD189 pKa = 4.3LMPKK193 pKa = 9.66VGVYY197 pKa = 8.98TSQSGSGSVDD207 pKa = 3.43VITSLDD213 pKa = 3.35TCAWTAVSDD222 pKa = 4.06NSWITIDD229 pKa = 3.89AGSNGTGNAAVSYY242 pKa = 10.31SVSEE246 pKa = 4.34NPNTSPRR253 pKa = 11.84IGILEE258 pKa = 3.66IAGRR262 pKa = 11.84KK263 pKa = 6.73FTVTQTGADD272 pKa = 3.66ATCSVSILPQQTTISSEE289 pKa = 4.06STEE292 pKa = 4.0GNIIVLTSPADD303 pKa = 4.04CAWSTTTVASSWIAISDD320 pKa = 4.01PEE322 pKa = 4.52SGSGTGDD329 pKa = 3.3SVITYY334 pKa = 9.56KK335 pKa = 11.02AEE337 pKa = 4.19ANADD341 pKa = 3.31TSDD344 pKa = 3.39RR345 pKa = 11.84FGVVMVNDD353 pKa = 3.76QPFMLTQKK361 pKa = 10.3GVCTYY366 pKa = 10.66DD367 pKa = 3.68LSPVVIDD374 pKa = 3.91IDD376 pKa = 3.91AAGSASEE383 pKa = 4.39TVAVTPSDD391 pKa = 5.56SICTWNAISDD401 pKa = 4.14EE402 pKa = 4.1DD403 pKa = 4.32WITVVSSSSSGTGTGTVEE421 pKa = 3.89FAVAANSTGAMRR433 pKa = 11.84TGKK436 pKa = 10.12VVIGGQNLLIVQLSEE451 pKa = 3.73AGKK454 pKa = 9.56PFDD457 pKa = 6.42DD458 pKa = 5.14IDD460 pKa = 4.97SEE462 pKa = 5.65DD463 pKa = 3.82DD464 pKa = 3.48TFQNAINAMKK474 pKa = 10.06AYY476 pKa = 10.14SITEE480 pKa = 4.03GCDD483 pKa = 3.15AQGNYY488 pKa = 9.83CPEE491 pKa = 4.59GYY493 pKa = 10.67VNRR496 pKa = 11.84AQMAVFVLKK505 pKa = 10.64AAGQAPSDD513 pKa = 3.77SDD515 pKa = 3.8TCTGAMFNDD524 pKa = 3.75VTTEE528 pKa = 3.77TLGDD532 pKa = 3.94YY533 pKa = 9.89FCKK536 pKa = 10.48AIEE539 pKa = 4.5KK540 pKa = 9.76FASLNITAGCGDD552 pKa = 4.37GNFCPDD558 pKa = 3.36GYY560 pKa = 8.94VTRR563 pKa = 11.84AQMAVFMLKK572 pKa = 10.72GLGQGPNSSDD582 pKa = 2.99KK583 pKa = 11.48CKK585 pKa = 10.08GTEE588 pKa = 4.24LNDD591 pKa = 2.96ITTDD595 pKa = 3.12KK596 pKa = 11.02VGEE599 pKa = 4.25YY600 pKa = 9.79FCKK603 pKa = 10.49AIEE606 pKa = 4.06KK607 pKa = 10.34FRR609 pKa = 11.84DD610 pKa = 3.33KK611 pKa = 11.19NITTGCSVDD620 pKa = 5.26DD621 pKa = 4.28PLTTEE626 pKa = 4.95NEE628 pKa = 3.78AAYY631 pKa = 10.74CPDD634 pKa = 4.06DD635 pKa = 3.75NVKK638 pKa = 10.11RR639 pKa = 11.84SQMALFLTKK648 pKa = 10.72GFLSDD653 pKa = 3.49KK654 pKa = 11.03
Molecular weight: 68.21 kDa
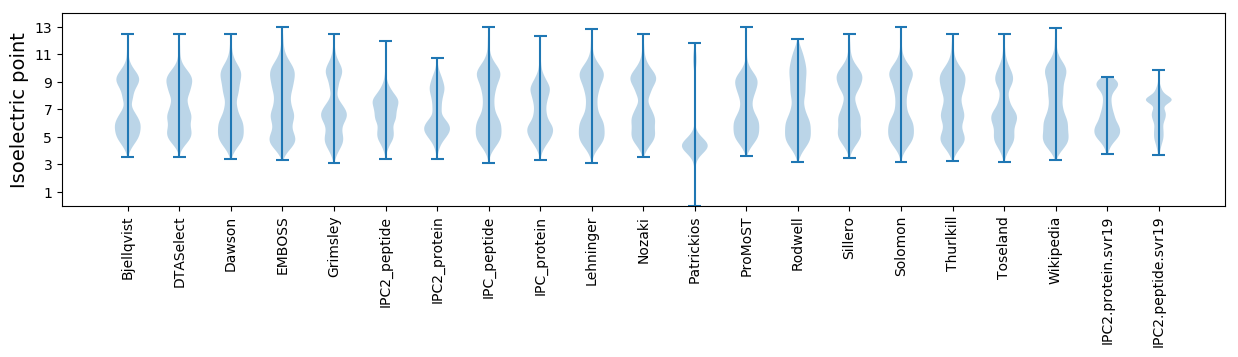
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F2J4J5|A0A0F2J4J5_9BACT ATP-dependent dethiobiotin synthetase BioD OS=Candidatus Magnetoovum chiemensis OX=1609970 GN=bioD PE=3 SV=1
MM1 pKa = 7.44IEE3 pKa = 4.01KK4 pKa = 10.37AKK6 pKa = 10.45RR7 pKa = 11.84PPKK10 pKa = 10.41FKK12 pKa = 9.8VRR14 pKa = 11.84QYY16 pKa = 11.05NRR18 pKa = 11.84CKK20 pKa = 9.97LCGRR24 pKa = 11.84ARR26 pKa = 11.84GYY28 pKa = 9.21YY29 pKa = 9.77RR30 pKa = 11.84KK31 pKa = 9.45FAICRR36 pKa = 11.84ICFRR40 pKa = 11.84TLSLEE45 pKa = 3.96GQIPGVTKK53 pKa = 10.75SSWW56 pKa = 2.87
MM1 pKa = 7.44IEE3 pKa = 4.01KK4 pKa = 10.37AKK6 pKa = 10.45RR7 pKa = 11.84PPKK10 pKa = 10.41FKK12 pKa = 9.8VRR14 pKa = 11.84QYY16 pKa = 11.05NRR18 pKa = 11.84CKK20 pKa = 9.97LCGRR24 pKa = 11.84ARR26 pKa = 11.84GYY28 pKa = 9.21YY29 pKa = 9.77RR30 pKa = 11.84KK31 pKa = 9.45FAICRR36 pKa = 11.84ICFRR40 pKa = 11.84TLSLEE45 pKa = 3.96GQIPGVTKK53 pKa = 10.75SSWW56 pKa = 2.87
Molecular weight: 6.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066827 |
27 |
2143 |
259.8 |
29.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.106 ± 0.042 | 1.195 ± 0.016 |
5.726 ± 0.035 | 6.657 ± 0.044 |
4.355 ± 0.029 | 5.96 ± 0.044 |
1.75 ± 0.019 | 8.898 ± 0.046 |
7.754 ± 0.051 | 9.712 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.019 | 5.214 ± 0.033 |
3.449 ± 0.026 | 2.985 ± 0.021 |
4.314 ± 0.027 | 6.687 ± 0.042 |
5.34 ± 0.029 | 5.896 ± 0.038 |
0.905 ± 0.015 | 3.684 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
