
Hubei sobemo-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
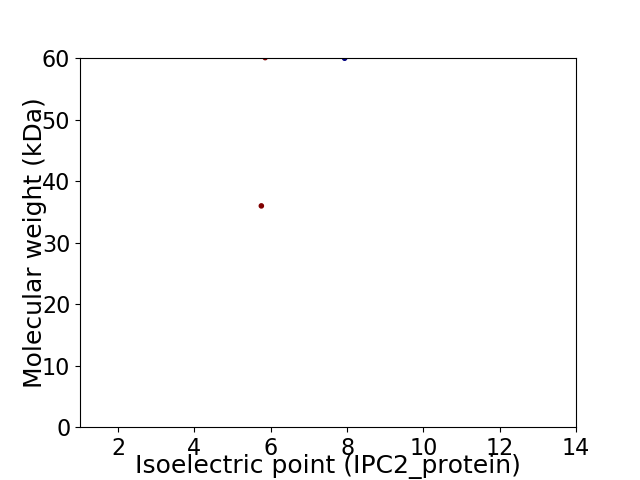
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KES0|A0A1L3KES0_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 11 OX=1923196 PE=4 SV=1
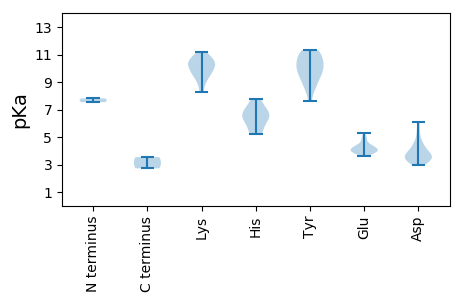
MM1 pKa = 7.53LKK3 pKa = 10.46NVVLEE8 pKa = 4.12RR9 pKa = 11.84MHH11 pKa = 7.2RR12 pKa = 11.84LINGADD18 pKa = 3.51VFDD21 pKa = 6.09DD22 pKa = 3.28IQVFIKK28 pKa = 10.43QEE30 pKa = 3.65PHH32 pKa = 6.54KK33 pKa = 10.45LRR35 pKa = 11.84KK36 pKa = 9.22IEE38 pKa = 3.86EE39 pKa = 4.06GRR41 pKa = 11.84FRR43 pKa = 11.84LISGVSFVDD52 pKa = 4.86GIIDD56 pKa = 4.23RR57 pKa = 11.84IVFGWLADD65 pKa = 3.49AAMNAIGQTPALLGWNPLFGGDD87 pKa = 3.46RR88 pKa = 11.84LLFSKK93 pKa = 10.31IQAPFFSLDD102 pKa = 3.01KK103 pKa = 11.15SLWDD107 pKa = 3.22WTVPAWLVDD116 pKa = 3.46LWLQFILGLYY126 pKa = 10.0HH127 pKa = 7.75DD128 pKa = 4.1MPEE131 pKa = 3.94WLEE134 pKa = 3.77LLIVKK139 pKa = 9.71RR140 pKa = 11.84FNILFAKK147 pKa = 10.27SRR149 pKa = 11.84FRR151 pKa = 11.84FPDD154 pKa = 3.42GCVIEE159 pKa = 4.51QKK161 pKa = 10.65FSGIMKK167 pKa = 9.7SGCFLTILLNSVGQYY182 pKa = 10.43LVDD185 pKa = 5.29RR186 pKa = 11.84IVSYY190 pKa = 11.09NLNIPTGEE198 pKa = 4.01ILCYY202 pKa = 10.58GDD204 pKa = 4.61DD205 pKa = 4.46TIQEE209 pKa = 4.17VPDD212 pKa = 3.5NVEE215 pKa = 4.01NYY217 pKa = 9.4ISAVRR222 pKa = 11.84ALGFLPKK229 pKa = 10.54DD230 pKa = 3.71PIVSNVAEE238 pKa = 4.09FVGFVMDD245 pKa = 3.73GRR247 pKa = 11.84KK248 pKa = 9.57VIPAYY253 pKa = 10.21YY254 pKa = 9.9KK255 pKa = 9.63KK256 pKa = 9.85HH257 pKa = 6.08LYY259 pKa = 9.39NLRR262 pKa = 11.84HH263 pKa = 6.39LDD265 pKa = 3.59EE266 pKa = 4.96KK267 pKa = 11.07VAEE270 pKa = 4.27ATLEE274 pKa = 4.0NYY276 pKa = 10.03QILYY280 pKa = 10.43ADD282 pKa = 4.21SEE284 pKa = 4.41EE285 pKa = 4.26LLPVIQAEE293 pKa = 4.26LTQRR297 pKa = 11.84NPTRR301 pKa = 11.84VIPRR305 pKa = 11.84VALQRR310 pKa = 11.84VVRR313 pKa = 11.84GG314 pKa = 3.54
MM1 pKa = 7.53LKK3 pKa = 10.46NVVLEE8 pKa = 4.12RR9 pKa = 11.84MHH11 pKa = 7.2RR12 pKa = 11.84LINGADD18 pKa = 3.51VFDD21 pKa = 6.09DD22 pKa = 3.28IQVFIKK28 pKa = 10.43QEE30 pKa = 3.65PHH32 pKa = 6.54KK33 pKa = 10.45LRR35 pKa = 11.84KK36 pKa = 9.22IEE38 pKa = 3.86EE39 pKa = 4.06GRR41 pKa = 11.84FRR43 pKa = 11.84LISGVSFVDD52 pKa = 4.86GIIDD56 pKa = 4.23RR57 pKa = 11.84IVFGWLADD65 pKa = 3.49AAMNAIGQTPALLGWNPLFGGDD87 pKa = 3.46RR88 pKa = 11.84LLFSKK93 pKa = 10.31IQAPFFSLDD102 pKa = 3.01KK103 pKa = 11.15SLWDD107 pKa = 3.22WTVPAWLVDD116 pKa = 3.46LWLQFILGLYY126 pKa = 10.0HH127 pKa = 7.75DD128 pKa = 4.1MPEE131 pKa = 3.94WLEE134 pKa = 3.77LLIVKK139 pKa = 9.71RR140 pKa = 11.84FNILFAKK147 pKa = 10.27SRR149 pKa = 11.84FRR151 pKa = 11.84FPDD154 pKa = 3.42GCVIEE159 pKa = 4.51QKK161 pKa = 10.65FSGIMKK167 pKa = 9.7SGCFLTILLNSVGQYY182 pKa = 10.43LVDD185 pKa = 5.29RR186 pKa = 11.84IVSYY190 pKa = 11.09NLNIPTGEE198 pKa = 4.01ILCYY202 pKa = 10.58GDD204 pKa = 4.61DD205 pKa = 4.46TIQEE209 pKa = 4.17VPDD212 pKa = 3.5NVEE215 pKa = 4.01NYY217 pKa = 9.4ISAVRR222 pKa = 11.84ALGFLPKK229 pKa = 10.54DD230 pKa = 3.71PIVSNVAEE238 pKa = 4.09FVGFVMDD245 pKa = 3.73GRR247 pKa = 11.84KK248 pKa = 9.57VIPAYY253 pKa = 10.21YY254 pKa = 9.9KK255 pKa = 9.63KK256 pKa = 9.85HH257 pKa = 6.08LYY259 pKa = 9.39NLRR262 pKa = 11.84HH263 pKa = 6.39LDD265 pKa = 3.59EE266 pKa = 4.96KK267 pKa = 11.07VAEE270 pKa = 4.27ATLEE274 pKa = 4.0NYY276 pKa = 10.03QILYY280 pKa = 10.43ADD282 pKa = 4.21SEE284 pKa = 4.41EE285 pKa = 4.26LLPVIQAEE293 pKa = 4.26LTQRR297 pKa = 11.84NPTRR301 pKa = 11.84VIPRR305 pKa = 11.84VALQRR310 pKa = 11.84VVRR313 pKa = 11.84GG314 pKa = 3.54
Molecular weight: 36.0 kDa
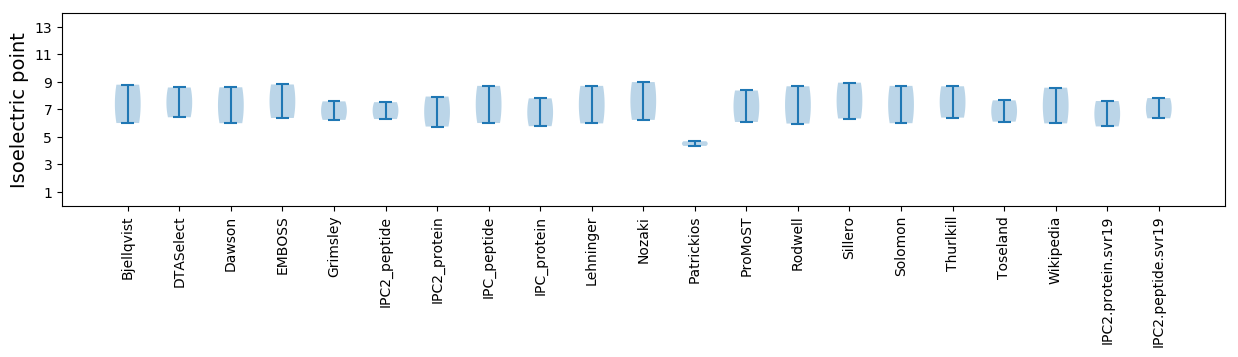
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KES0|A0A1L3KES0_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 11 OX=1923196 PE=4 SV=1
MM1 pKa = 7.81ALRR4 pKa = 11.84FVCEE8 pKa = 3.73VLFWILFMVFKK19 pKa = 10.45VLWRR23 pKa = 11.84FEE25 pKa = 4.28LGRR28 pKa = 11.84CILAFLSVFGLVLFWNRR45 pKa = 11.84VIDD48 pKa = 4.29LLWLVWVILSGLWRR62 pKa = 11.84VKK64 pKa = 9.9PVAVAPEE71 pKa = 4.19PVSPASMLLWWFIIVALLGILMLTYY96 pKa = 11.19LLFLWVFVKK105 pKa = 10.57IKK107 pKa = 10.27SAIVGKK113 pKa = 10.02QIEE116 pKa = 4.37KK117 pKa = 10.34FAFEE121 pKa = 5.0RR122 pKa = 11.84YY123 pKa = 8.54QEE125 pKa = 4.19GSPYY129 pKa = 10.8LSAKK133 pKa = 10.17CPDD136 pKa = 3.69VQCEE140 pKa = 3.97VWVSYY145 pKa = 11.2DD146 pKa = 3.78GDD148 pKa = 3.8WLKK151 pKa = 10.99SGQAFRR157 pKa = 11.84VGDD160 pKa = 3.66KK161 pKa = 10.84LITAGHH167 pKa = 5.19VVRR170 pKa = 11.84EE171 pKa = 3.96ARR173 pKa = 11.84QAKK176 pKa = 8.51FVSGAGEE183 pKa = 4.07VFVDD187 pKa = 3.34VDD189 pKa = 3.81SFAEE193 pKa = 3.95IEE195 pKa = 4.08GDD197 pKa = 3.91VVFVRR202 pKa = 11.84LSNLDD207 pKa = 3.12ITTLALKK214 pKa = 9.94SAKK217 pKa = 9.07FANVGCDD224 pKa = 3.03EE225 pKa = 4.57GKK227 pKa = 8.85GTFCTVQAFGRR238 pKa = 11.84MSMGMVEE245 pKa = 4.41SFPAFGFCKK254 pKa = 9.51YY255 pKa = 9.07TGSTDD260 pKa = 3.12HH261 pKa = 6.65GFSGAPYY268 pKa = 9.59LVNRR272 pKa = 11.84TVFGVHH278 pKa = 6.77IGTQGVNLGYY288 pKa = 10.6EE289 pKa = 4.32GAYY292 pKa = 9.89VAACLRR298 pKa = 11.84SRR300 pKa = 11.84NEE302 pKa = 3.76DD303 pKa = 3.39SEE305 pKa = 4.86EE306 pKa = 4.01YY307 pKa = 11.05LMKK310 pKa = 10.27QVKK313 pKa = 9.58KK314 pKa = 10.12GRR316 pKa = 11.84GVARR320 pKa = 11.84QSPYY324 pKa = 11.3DD325 pKa = 3.81PDD327 pKa = 3.35EE328 pKa = 4.09VQIQVGGKK336 pKa = 8.41YY337 pKa = 7.6FTMSRR342 pKa = 11.84DD343 pKa = 3.48AYY345 pKa = 9.57EE346 pKa = 3.96QAQEE350 pKa = 3.77EE351 pKa = 4.53MEE353 pKa = 4.04RR354 pKa = 11.84DD355 pKa = 3.87FEE357 pKa = 5.28RR358 pKa = 11.84KK359 pKa = 9.38FDD361 pKa = 4.53DD362 pKa = 5.33DD363 pKa = 3.74SGRR366 pKa = 11.84WGGGKK371 pKa = 9.84NYY373 pKa = 8.57HH374 pKa = 5.44TGKK377 pKa = 9.91NRR379 pKa = 11.84GGRR382 pKa = 11.84LVRR385 pKa = 11.84EE386 pKa = 4.26SALEE390 pKa = 3.79RR391 pKa = 11.84SEE393 pKa = 4.28PLPTFDD399 pKa = 5.93DD400 pKa = 4.1VQSEE404 pKa = 4.28PKK406 pKa = 10.14NVGRR410 pKa = 11.84IEE412 pKa = 4.01NAGVSISPKK421 pKa = 10.29KK422 pKa = 8.27PASWYY427 pKa = 8.77EE428 pKa = 3.82DD429 pKa = 2.99QEE431 pKa = 4.74VFVRR435 pKa = 11.84KK436 pKa = 9.35PNIQIGPSTSTPSTTIPVTVRR457 pKa = 11.84PDD459 pKa = 3.26MVLPEE464 pKa = 4.73SIAVPRR470 pKa = 11.84RR471 pKa = 11.84NPSSPTSKK479 pKa = 10.15NAKK482 pKa = 8.99KK483 pKa = 9.61QAKK486 pKa = 9.12ALQRR490 pKa = 11.84EE491 pKa = 4.4KK492 pKa = 10.98LRR494 pKa = 11.84SEE496 pKa = 5.07LLTQLGKK503 pKa = 10.72ALGIGDD509 pKa = 4.54RR510 pKa = 11.84EE511 pKa = 3.89ILKK514 pKa = 10.86KK515 pKa = 10.73NFSSSSNEE523 pKa = 3.84SLKK526 pKa = 10.84EE527 pKa = 3.69LLGSVTSKK535 pKa = 10.74PVV537 pKa = 2.72
MM1 pKa = 7.81ALRR4 pKa = 11.84FVCEE8 pKa = 3.73VLFWILFMVFKK19 pKa = 10.45VLWRR23 pKa = 11.84FEE25 pKa = 4.28LGRR28 pKa = 11.84CILAFLSVFGLVLFWNRR45 pKa = 11.84VIDD48 pKa = 4.29LLWLVWVILSGLWRR62 pKa = 11.84VKK64 pKa = 9.9PVAVAPEE71 pKa = 4.19PVSPASMLLWWFIIVALLGILMLTYY96 pKa = 11.19LLFLWVFVKK105 pKa = 10.57IKK107 pKa = 10.27SAIVGKK113 pKa = 10.02QIEE116 pKa = 4.37KK117 pKa = 10.34FAFEE121 pKa = 5.0RR122 pKa = 11.84YY123 pKa = 8.54QEE125 pKa = 4.19GSPYY129 pKa = 10.8LSAKK133 pKa = 10.17CPDD136 pKa = 3.69VQCEE140 pKa = 3.97VWVSYY145 pKa = 11.2DD146 pKa = 3.78GDD148 pKa = 3.8WLKK151 pKa = 10.99SGQAFRR157 pKa = 11.84VGDD160 pKa = 3.66KK161 pKa = 10.84LITAGHH167 pKa = 5.19VVRR170 pKa = 11.84EE171 pKa = 3.96ARR173 pKa = 11.84QAKK176 pKa = 8.51FVSGAGEE183 pKa = 4.07VFVDD187 pKa = 3.34VDD189 pKa = 3.81SFAEE193 pKa = 3.95IEE195 pKa = 4.08GDD197 pKa = 3.91VVFVRR202 pKa = 11.84LSNLDD207 pKa = 3.12ITTLALKK214 pKa = 9.94SAKK217 pKa = 9.07FANVGCDD224 pKa = 3.03EE225 pKa = 4.57GKK227 pKa = 8.85GTFCTVQAFGRR238 pKa = 11.84MSMGMVEE245 pKa = 4.41SFPAFGFCKK254 pKa = 9.51YY255 pKa = 9.07TGSTDD260 pKa = 3.12HH261 pKa = 6.65GFSGAPYY268 pKa = 9.59LVNRR272 pKa = 11.84TVFGVHH278 pKa = 6.77IGTQGVNLGYY288 pKa = 10.6EE289 pKa = 4.32GAYY292 pKa = 9.89VAACLRR298 pKa = 11.84SRR300 pKa = 11.84NEE302 pKa = 3.76DD303 pKa = 3.39SEE305 pKa = 4.86EE306 pKa = 4.01YY307 pKa = 11.05LMKK310 pKa = 10.27QVKK313 pKa = 9.58KK314 pKa = 10.12GRR316 pKa = 11.84GVARR320 pKa = 11.84QSPYY324 pKa = 11.3DD325 pKa = 3.81PDD327 pKa = 3.35EE328 pKa = 4.09VQIQVGGKK336 pKa = 8.41YY337 pKa = 7.6FTMSRR342 pKa = 11.84DD343 pKa = 3.48AYY345 pKa = 9.57EE346 pKa = 3.96QAQEE350 pKa = 3.77EE351 pKa = 4.53MEE353 pKa = 4.04RR354 pKa = 11.84DD355 pKa = 3.87FEE357 pKa = 5.28RR358 pKa = 11.84KK359 pKa = 9.38FDD361 pKa = 4.53DD362 pKa = 5.33DD363 pKa = 3.74SGRR366 pKa = 11.84WGGGKK371 pKa = 9.84NYY373 pKa = 8.57HH374 pKa = 5.44TGKK377 pKa = 9.91NRR379 pKa = 11.84GGRR382 pKa = 11.84LVRR385 pKa = 11.84EE386 pKa = 4.26SALEE390 pKa = 3.79RR391 pKa = 11.84SEE393 pKa = 4.28PLPTFDD399 pKa = 5.93DD400 pKa = 4.1VQSEE404 pKa = 4.28PKK406 pKa = 10.14NVGRR410 pKa = 11.84IEE412 pKa = 4.01NAGVSISPKK421 pKa = 10.29KK422 pKa = 8.27PASWYY427 pKa = 8.77EE428 pKa = 3.82DD429 pKa = 2.99QEE431 pKa = 4.74VFVRR435 pKa = 11.84KK436 pKa = 9.35PNIQIGPSTSTPSTTIPVTVRR457 pKa = 11.84PDD459 pKa = 3.26MVLPEE464 pKa = 4.73SIAVPRR470 pKa = 11.84RR471 pKa = 11.84NPSSPTSKK479 pKa = 10.15NAKK482 pKa = 8.99KK483 pKa = 9.61QAKK486 pKa = 9.12ALQRR490 pKa = 11.84EE491 pKa = 4.4KK492 pKa = 10.98LRR494 pKa = 11.84SEE496 pKa = 5.07LLTQLGKK503 pKa = 10.72ALGIGDD509 pKa = 4.54RR510 pKa = 11.84EE511 pKa = 3.89ILKK514 pKa = 10.86KK515 pKa = 10.73NFSSSSNEE523 pKa = 3.84SLKK526 pKa = 10.84EE527 pKa = 3.69LLGSVTSKK535 pKa = 10.74PVV537 pKa = 2.72
Molecular weight: 60.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
851 |
314 |
537 |
425.5 |
48.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.228 ± 0.282 | 1.293 ± 0.192 |
5.17 ± 0.683 | 6.228 ± 0.464 |
5.993 ± 0.033 | 7.638 ± 0.723 |
1.058 ± 0.305 | 5.758 ± 1.437 |
5.875 ± 0.626 | 10.223 ± 1.252 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.998 ± 0.049 | 3.408 ± 0.599 |
4.7 ± 0.044 | 3.643 ± 0.102 |
5.993 ± 0.033 | 6.58 ± 1.39 |
3.408 ± 0.49 | 9.636 ± 0.409 |
2.35 ± 0.069 | 2.82 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
