
Kineothrix alysoides
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Kineothrix
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
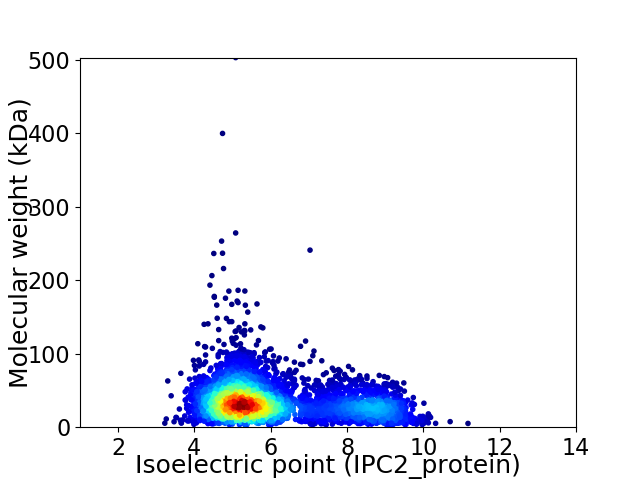
Virtual 2D-PAGE plot for 4121 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V2QB24|A0A4V2QB24_9FIRM N-acetylmuramoyl-L-alanine amidase OS=Kineothrix alysoides OX=1469948 GN=EDD76_1183 PE=4 SV=1
MM1 pKa = 7.57SKK3 pKa = 9.49TNKK6 pKa = 9.57FLALCAAFVGLGIILYY22 pKa = 8.73ITGFFLGEE30 pKa = 4.21SVTGIGISNDD40 pKa = 2.87GFQVYY45 pKa = 8.48SWNQTKK51 pKa = 10.7LNEE54 pKa = 4.01TDD56 pKa = 3.9FIEE59 pKa = 4.32MTDD62 pKa = 3.26IEE64 pKa = 5.37LDD66 pKa = 3.21AFQNIDD72 pKa = 3.56MKK74 pKa = 11.16VSYY77 pKa = 11.23ANVEE81 pKa = 3.97IKK83 pKa = 10.62EE84 pKa = 3.97SDD86 pKa = 3.3RR87 pKa = 11.84FKK89 pKa = 10.81ISYY92 pKa = 8.01KK93 pKa = 9.26TAQNHH98 pKa = 5.45PLSYY102 pKa = 9.94EE103 pKa = 3.92IKK105 pKa = 10.56DD106 pKa = 3.65GTLHH110 pKa = 6.21ITQANNAAGSSSFHH124 pKa = 7.32YY125 pKa = 9.94MFLGFSGIQGGDD137 pKa = 2.76IRR139 pKa = 11.84EE140 pKa = 4.24NEE142 pKa = 4.39SVTVYY147 pKa = 10.62IPGNVEE153 pKa = 3.94LNSISVNNDD162 pKa = 2.92CGNTNLSSLNASSVSISADD181 pKa = 3.37YY182 pKa = 11.55GDD184 pKa = 4.32VNLTGMNAPCIALEE198 pKa = 4.23VNTGYY203 pKa = 11.38LNLDD207 pKa = 3.79NIIADD212 pKa = 4.16TLTVTTDD219 pKa = 3.27YY220 pKa = 11.94GNITLNTIDD229 pKa = 4.03TSTAALKK236 pKa = 11.14VNTGNVNLDD245 pKa = 3.82SIVADD250 pKa = 4.0TLTVTADD257 pKa = 3.33YY258 pKa = 11.31GDD260 pKa = 4.81AGLSTIDD267 pKa = 3.75ASSVTLKK274 pKa = 10.67MNTGNVNIRR283 pKa = 11.84DD284 pKa = 3.64IDD286 pKa = 3.72SGIIEE291 pKa = 4.14TAIDD295 pKa = 3.66YY296 pKa = 10.77GGLQLSSAEE305 pKa = 3.9TDD307 pKa = 3.5SFSVEE312 pKa = 3.87INTGSLEE319 pKa = 4.18ITDD322 pKa = 3.5MTANTFAADD331 pKa = 3.31IEE333 pKa = 4.43YY334 pKa = 11.02GEE336 pKa = 4.5TNAEE340 pKa = 4.28RR341 pKa = 11.84ITISGDD347 pKa = 2.91ADD349 pKa = 2.93ITMGTGSLHH358 pKa = 6.25MKK360 pKa = 10.06DD361 pKa = 5.17ASLQTLTADD370 pKa = 3.43NEE372 pKa = 4.38YY373 pKa = 11.16GDD375 pKa = 3.8VSLEE379 pKa = 4.01FSSPATDD386 pKa = 3.4YY387 pKa = 11.39SYY389 pKa = 11.76DD390 pKa = 3.37LFTEE394 pKa = 4.01YY395 pKa = 9.61GTIKK399 pKa = 10.82VEE401 pKa = 4.09GQDD404 pKa = 2.99MGEE407 pKa = 4.12KK408 pKa = 9.98YY409 pKa = 10.21KK410 pKa = 10.49PLSSSDD416 pKa = 3.03IGKK419 pKa = 9.4HH420 pKa = 4.0ITVSCEE426 pKa = 3.71SGNIDD431 pKa = 4.29IKK433 pKa = 11.33GLL435 pKa = 3.62
MM1 pKa = 7.57SKK3 pKa = 9.49TNKK6 pKa = 9.57FLALCAAFVGLGIILYY22 pKa = 8.73ITGFFLGEE30 pKa = 4.21SVTGIGISNDD40 pKa = 2.87GFQVYY45 pKa = 8.48SWNQTKK51 pKa = 10.7LNEE54 pKa = 4.01TDD56 pKa = 3.9FIEE59 pKa = 4.32MTDD62 pKa = 3.26IEE64 pKa = 5.37LDD66 pKa = 3.21AFQNIDD72 pKa = 3.56MKK74 pKa = 11.16VSYY77 pKa = 11.23ANVEE81 pKa = 3.97IKK83 pKa = 10.62EE84 pKa = 3.97SDD86 pKa = 3.3RR87 pKa = 11.84FKK89 pKa = 10.81ISYY92 pKa = 8.01KK93 pKa = 9.26TAQNHH98 pKa = 5.45PLSYY102 pKa = 9.94EE103 pKa = 3.92IKK105 pKa = 10.56DD106 pKa = 3.65GTLHH110 pKa = 6.21ITQANNAAGSSSFHH124 pKa = 7.32YY125 pKa = 9.94MFLGFSGIQGGDD137 pKa = 2.76IRR139 pKa = 11.84EE140 pKa = 4.24NEE142 pKa = 4.39SVTVYY147 pKa = 10.62IPGNVEE153 pKa = 3.94LNSISVNNDD162 pKa = 2.92CGNTNLSSLNASSVSISADD181 pKa = 3.37YY182 pKa = 11.55GDD184 pKa = 4.32VNLTGMNAPCIALEE198 pKa = 4.23VNTGYY203 pKa = 11.38LNLDD207 pKa = 3.79NIIADD212 pKa = 4.16TLTVTTDD219 pKa = 3.27YY220 pKa = 11.94GNITLNTIDD229 pKa = 4.03TSTAALKK236 pKa = 11.14VNTGNVNLDD245 pKa = 3.82SIVADD250 pKa = 4.0TLTVTADD257 pKa = 3.33YY258 pKa = 11.31GDD260 pKa = 4.81AGLSTIDD267 pKa = 3.75ASSVTLKK274 pKa = 10.67MNTGNVNIRR283 pKa = 11.84DD284 pKa = 3.64IDD286 pKa = 3.72SGIIEE291 pKa = 4.14TAIDD295 pKa = 3.66YY296 pKa = 10.77GGLQLSSAEE305 pKa = 3.9TDD307 pKa = 3.5SFSVEE312 pKa = 3.87INTGSLEE319 pKa = 4.18ITDD322 pKa = 3.5MTANTFAADD331 pKa = 3.31IEE333 pKa = 4.43YY334 pKa = 11.02GEE336 pKa = 4.5TNAEE340 pKa = 4.28RR341 pKa = 11.84ITISGDD347 pKa = 2.91ADD349 pKa = 2.93ITMGTGSLHH358 pKa = 6.25MKK360 pKa = 10.06DD361 pKa = 5.17ASLQTLTADD370 pKa = 3.43NEE372 pKa = 4.38YY373 pKa = 11.16GDD375 pKa = 3.8VSLEE379 pKa = 4.01FSSPATDD386 pKa = 3.4YY387 pKa = 11.39SYY389 pKa = 11.76DD390 pKa = 3.37LFTEE394 pKa = 4.01YY395 pKa = 9.61GTIKK399 pKa = 10.82VEE401 pKa = 4.09GQDD404 pKa = 2.99MGEE407 pKa = 4.12KK408 pKa = 9.98YY409 pKa = 10.21KK410 pKa = 10.49PLSSSDD416 pKa = 3.03IGKK419 pKa = 9.4HH420 pKa = 4.0ITVSCEE426 pKa = 3.71SGNIDD431 pKa = 4.29IKK433 pKa = 11.33GLL435 pKa = 3.62
Molecular weight: 46.54 kDa
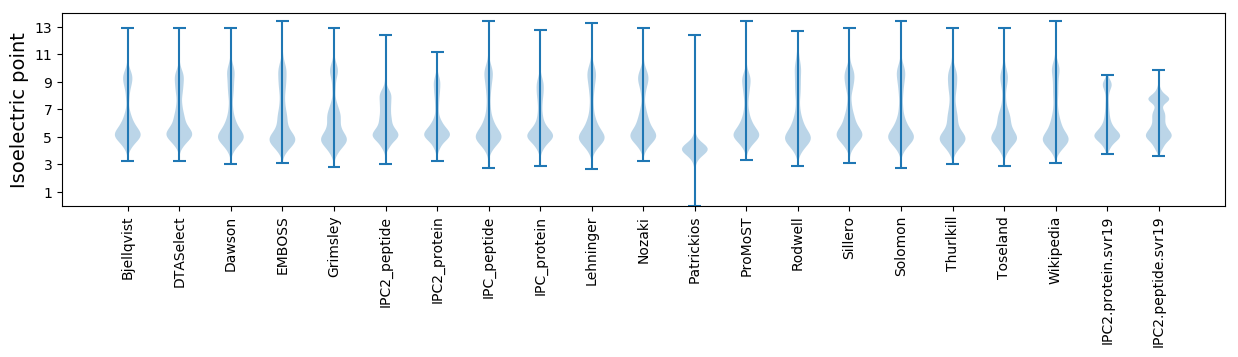
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1QV28|A0A4R1QV28_9FIRM Flagellar hook-associated protein 1 OS=Kineothrix alysoides OX=1469948 GN=flgK PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.55NRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.77KK14 pKa = 7.79VHH16 pKa = 5.69GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.55NRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.77KK14 pKa = 7.79VHH16 pKa = 5.69GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1351134 |
29 |
4427 |
327.9 |
36.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.28 ± 0.044 | 1.376 ± 0.017 |
5.43 ± 0.028 | 7.787 ± 0.049 |
4.262 ± 0.03 | 7.172 ± 0.039 |
1.621 ± 0.018 | 7.926 ± 0.035 |
6.876 ± 0.033 | 8.904 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.122 ± 0.019 | 4.59 ± 0.03 |
3.106 ± 0.021 | 3.063 ± 0.022 |
4.156 ± 0.03 | 6.112 ± 0.036 |
5.265 ± 0.045 | 6.67 ± 0.032 |
0.939 ± 0.014 | 4.342 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
