
Bittarella massiliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Bittarella
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
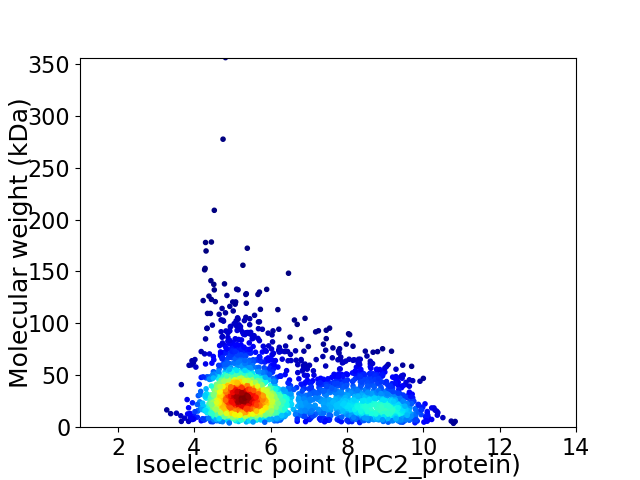
Virtual 2D-PAGE plot for 2773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B1UCL2|A0A6B1UCL2_9FIRM DUF4314 domain-containing protein OS=Bittarella massiliensis OX=1720313 GN=GT747_03940 PE=4 SV=1
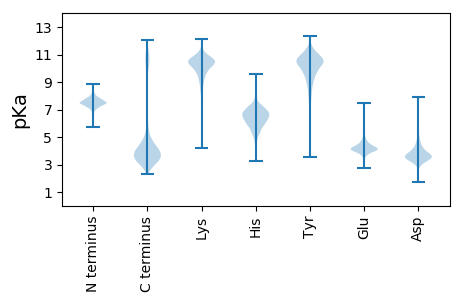
MM1 pKa = 7.52KK2 pKa = 10.26LKK4 pKa = 10.76SLAALMMAGVMALGLCACTNSGTPDD29 pKa = 3.63DD30 pKa = 5.12SSTSDD35 pKa = 2.89TSEE38 pKa = 3.81AANAEE43 pKa = 4.3PVSITTTEE51 pKa = 3.63SWDD54 pKa = 3.71FSTGFYY60 pKa = 9.45PALSAGSSNGTMGFTYY76 pKa = 10.27YY77 pKa = 11.03ARR79 pKa = 11.84NCYY82 pKa = 8.0DD83 pKa = 3.33TLVVRR88 pKa = 11.84EE89 pKa = 4.01GSEE92 pKa = 4.25FKK94 pKa = 11.04GGLAEE99 pKa = 4.12TWDD102 pKa = 3.66ISDD105 pKa = 5.5DD106 pKa = 3.48GLTYY110 pKa = 10.38TFHH113 pKa = 7.49LKK115 pKa = 10.6EE116 pKa = 4.12GVKK119 pKa = 10.64FSDD122 pKa = 3.74GADD125 pKa = 3.69FNAEE129 pKa = 3.81AVKK132 pKa = 10.07TSIEE136 pKa = 3.94AAFSNMGQSIAQFGKK151 pKa = 10.11IGVLTEE157 pKa = 4.25SIEE160 pKa = 4.35VIDD163 pKa = 4.86EE164 pKa = 4.26YY165 pKa = 11.24TVALHH170 pKa = 6.38LTTPYY175 pKa = 10.81YY176 pKa = 11.0GVLSDD181 pKa = 5.78LAMCNPMGIVSPNAFNEE198 pKa = 3.92DD199 pKa = 3.51LTTKK203 pKa = 10.45EE204 pKa = 4.16EE205 pKa = 5.08LMTQTMGTGPYY216 pKa = 8.77MYY218 pKa = 10.45AGDD221 pKa = 4.22SDD223 pKa = 3.83GTSYY227 pKa = 10.73TFVRR231 pKa = 11.84NPNYY235 pKa = 9.58WGEE238 pKa = 4.07EE239 pKa = 3.81PDD241 pKa = 3.37VDD243 pKa = 3.78EE244 pKa = 4.76FTVKK248 pKa = 10.57VIADD252 pKa = 3.66PEE254 pKa = 4.58SAVLALQSGEE264 pKa = 4.05VDD266 pKa = 3.88LLAGTSRR273 pKa = 11.84LSFAGYY279 pKa = 8.69TEE281 pKa = 4.99LSSADD286 pKa = 4.0GIGTTVDD293 pKa = 3.66DD294 pKa = 4.18SVSNSRR300 pKa = 11.84FIGFNTQKK308 pKa = 11.19APFNDD313 pKa = 3.39AAVRR317 pKa = 11.84QAVAYY322 pKa = 10.01AIDD325 pKa = 4.0KK326 pKa = 7.58EE327 pKa = 4.85TISSNVFSGLEE338 pKa = 3.88TASTYY343 pKa = 11.35LLDD346 pKa = 4.06TSLPYY351 pKa = 10.52CDD353 pKa = 5.52VEE355 pKa = 4.56TTTYY359 pKa = 10.89SYY361 pKa = 11.4DD362 pKa = 3.5PEE364 pKa = 4.54KK365 pKa = 10.72AAEE368 pKa = 4.12LLEE371 pKa = 4.36SAGWVDD377 pKa = 3.77SDD379 pKa = 4.35GDD381 pKa = 4.11GIRR384 pKa = 11.84EE385 pKa = 3.81KK386 pKa = 10.95DD387 pKa = 3.42GTKK390 pKa = 10.66LSVTLDD396 pKa = 3.6YY397 pKa = 10.22ITNQGTLDD405 pKa = 3.82DD406 pKa = 4.48AALVIAQEE414 pKa = 4.0LGEE417 pKa = 4.34VGFEE421 pKa = 3.96VTPRR425 pKa = 11.84GADD428 pKa = 2.94NMTWFTQVSTDD439 pKa = 3.89FDD441 pKa = 4.83FSLHH445 pKa = 4.44TTYY448 pKa = 10.94GGYY451 pKa = 10.22YY452 pKa = 10.45DD453 pKa = 5.2PFLTMTNMNPEE464 pKa = 4.14MMSDD468 pKa = 4.24PILWQVALTMEE479 pKa = 4.56NGTNIIKK486 pKa = 10.36EE487 pKa = 4.15LDD489 pKa = 3.43STADD493 pKa = 3.56LDD495 pKa = 4.47RR496 pKa = 11.84VQEE499 pKa = 4.13IYY501 pKa = 11.37NEE503 pKa = 4.2VLTFTADD510 pKa = 3.32QAVLVPFTSAHH521 pKa = 5.46QYY523 pKa = 10.75AAFNSDD529 pKa = 3.48KK530 pKa = 10.85IEE532 pKa = 4.19SFSFGSDD539 pKa = 3.01QLFIEE544 pKa = 4.46IANVKK549 pKa = 10.22AKK551 pKa = 10.66
MM1 pKa = 7.52KK2 pKa = 10.26LKK4 pKa = 10.76SLAALMMAGVMALGLCACTNSGTPDD29 pKa = 3.63DD30 pKa = 5.12SSTSDD35 pKa = 2.89TSEE38 pKa = 3.81AANAEE43 pKa = 4.3PVSITTTEE51 pKa = 3.63SWDD54 pKa = 3.71FSTGFYY60 pKa = 9.45PALSAGSSNGTMGFTYY76 pKa = 10.27YY77 pKa = 11.03ARR79 pKa = 11.84NCYY82 pKa = 8.0DD83 pKa = 3.33TLVVRR88 pKa = 11.84EE89 pKa = 4.01GSEE92 pKa = 4.25FKK94 pKa = 11.04GGLAEE99 pKa = 4.12TWDD102 pKa = 3.66ISDD105 pKa = 5.5DD106 pKa = 3.48GLTYY110 pKa = 10.38TFHH113 pKa = 7.49LKK115 pKa = 10.6EE116 pKa = 4.12GVKK119 pKa = 10.64FSDD122 pKa = 3.74GADD125 pKa = 3.69FNAEE129 pKa = 3.81AVKK132 pKa = 10.07TSIEE136 pKa = 3.94AAFSNMGQSIAQFGKK151 pKa = 10.11IGVLTEE157 pKa = 4.25SIEE160 pKa = 4.35VIDD163 pKa = 4.86EE164 pKa = 4.26YY165 pKa = 11.24TVALHH170 pKa = 6.38LTTPYY175 pKa = 10.81YY176 pKa = 11.0GVLSDD181 pKa = 5.78LAMCNPMGIVSPNAFNEE198 pKa = 3.92DD199 pKa = 3.51LTTKK203 pKa = 10.45EE204 pKa = 4.16EE205 pKa = 5.08LMTQTMGTGPYY216 pKa = 8.77MYY218 pKa = 10.45AGDD221 pKa = 4.22SDD223 pKa = 3.83GTSYY227 pKa = 10.73TFVRR231 pKa = 11.84NPNYY235 pKa = 9.58WGEE238 pKa = 4.07EE239 pKa = 3.81PDD241 pKa = 3.37VDD243 pKa = 3.78EE244 pKa = 4.76FTVKK248 pKa = 10.57VIADD252 pKa = 3.66PEE254 pKa = 4.58SAVLALQSGEE264 pKa = 4.05VDD266 pKa = 3.88LLAGTSRR273 pKa = 11.84LSFAGYY279 pKa = 8.69TEE281 pKa = 4.99LSSADD286 pKa = 4.0GIGTTVDD293 pKa = 3.66DD294 pKa = 4.18SVSNSRR300 pKa = 11.84FIGFNTQKK308 pKa = 11.19APFNDD313 pKa = 3.39AAVRR317 pKa = 11.84QAVAYY322 pKa = 10.01AIDD325 pKa = 4.0KK326 pKa = 7.58EE327 pKa = 4.85TISSNVFSGLEE338 pKa = 3.88TASTYY343 pKa = 11.35LLDD346 pKa = 4.06TSLPYY351 pKa = 10.52CDD353 pKa = 5.52VEE355 pKa = 4.56TTTYY359 pKa = 10.89SYY361 pKa = 11.4DD362 pKa = 3.5PEE364 pKa = 4.54KK365 pKa = 10.72AAEE368 pKa = 4.12LLEE371 pKa = 4.36SAGWVDD377 pKa = 3.77SDD379 pKa = 4.35GDD381 pKa = 4.11GIRR384 pKa = 11.84EE385 pKa = 3.81KK386 pKa = 10.95DD387 pKa = 3.42GTKK390 pKa = 10.66LSVTLDD396 pKa = 3.6YY397 pKa = 10.22ITNQGTLDD405 pKa = 3.82DD406 pKa = 4.48AALVIAQEE414 pKa = 4.0LGEE417 pKa = 4.34VGFEE421 pKa = 3.96VTPRR425 pKa = 11.84GADD428 pKa = 2.94NMTWFTQVSTDD439 pKa = 3.89FDD441 pKa = 4.83FSLHH445 pKa = 4.44TTYY448 pKa = 10.94GGYY451 pKa = 10.22YY452 pKa = 10.45DD453 pKa = 5.2PFLTMTNMNPEE464 pKa = 4.14MMSDD468 pKa = 4.24PILWQVALTMEE479 pKa = 4.56NGTNIIKK486 pKa = 10.36EE487 pKa = 4.15LDD489 pKa = 3.43STADD493 pKa = 3.56LDD495 pKa = 4.47RR496 pKa = 11.84VQEE499 pKa = 4.13IYY501 pKa = 11.37NEE503 pKa = 4.2VLTFTADD510 pKa = 3.32QAVLVPFTSAHH521 pKa = 5.46QYY523 pKa = 10.75AAFNSDD529 pKa = 3.48KK530 pKa = 10.85IEE532 pKa = 4.19SFSFGSDD539 pKa = 3.01QLFIEE544 pKa = 4.46IANVKK549 pKa = 10.22AKK551 pKa = 10.66
Molecular weight: 59.62 kDa
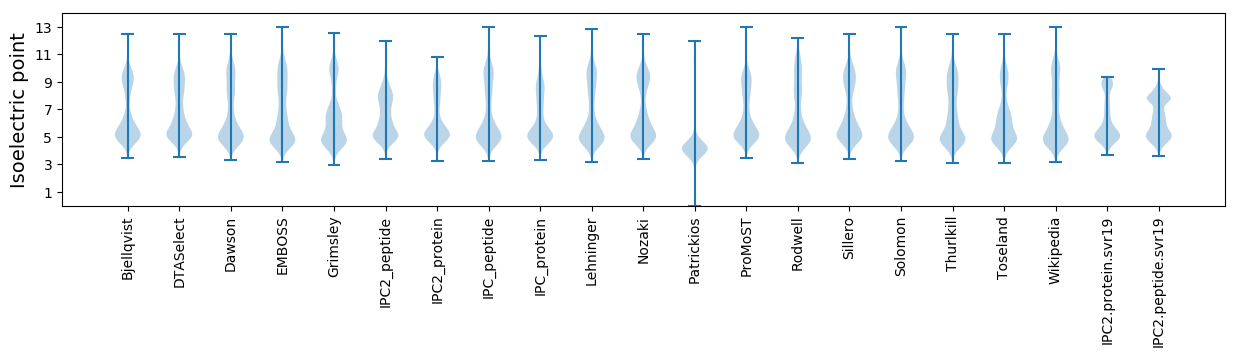
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B1UDH2|A0A6B1UDH2_9FIRM Transcriptional repressor NrdR OS=Bittarella massiliensis OX=1720313 GN=nrdR PE=3 SV=1
MM1 pKa = 7.23GRR3 pKa = 11.84ARR5 pKa = 11.84PVGEE9 pKa = 3.49KK10 pKa = 9.91RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 3.97KK15 pKa = 11.24EE16 pKa = 3.74MVAEE20 pKa = 5.12MIALYY25 pKa = 10.54CRR27 pKa = 11.84GRR29 pKa = 11.84HH30 pKa = 4.9GSRR33 pKa = 11.84GTLCPQCAEE42 pKa = 4.06LAAYY46 pKa = 9.0AAARR50 pKa = 11.84SDD52 pKa = 3.15RR53 pKa = 11.84CPFMEE58 pKa = 4.7EE59 pKa = 3.64KK60 pKa = 9.95TFCSNCRR67 pKa = 11.84VHH69 pKa = 7.4CYY71 pKa = 9.61RR72 pKa = 11.84PEE74 pKa = 3.33MRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84IRR80 pKa = 11.84EE81 pKa = 3.82VMRR84 pKa = 11.84YY85 pKa = 9.05AGPRR89 pKa = 11.84MLFHH93 pKa = 6.99HH94 pKa = 6.96PVAAIRR100 pKa = 11.84HH101 pKa = 5.39ALLTLRR107 pKa = 11.84EE108 pKa = 4.14KK109 pKa = 10.79KK110 pKa = 10.28RR111 pKa = 11.84LEE113 pKa = 4.04GEE115 pKa = 4.01RR116 pKa = 3.84
MM1 pKa = 7.23GRR3 pKa = 11.84ARR5 pKa = 11.84PVGEE9 pKa = 3.49KK10 pKa = 9.91RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 3.97KK15 pKa = 11.24EE16 pKa = 3.74MVAEE20 pKa = 5.12MIALYY25 pKa = 10.54CRR27 pKa = 11.84GRR29 pKa = 11.84HH30 pKa = 4.9GSRR33 pKa = 11.84GTLCPQCAEE42 pKa = 4.06LAAYY46 pKa = 9.0AAARR50 pKa = 11.84SDD52 pKa = 3.15RR53 pKa = 11.84CPFMEE58 pKa = 4.7EE59 pKa = 3.64KK60 pKa = 9.95TFCSNCRR67 pKa = 11.84VHH69 pKa = 7.4CYY71 pKa = 9.61RR72 pKa = 11.84PEE74 pKa = 3.33MRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84IRR80 pKa = 11.84EE81 pKa = 3.82VMRR84 pKa = 11.84YY85 pKa = 9.05AGPRR89 pKa = 11.84MLFHH93 pKa = 6.99HH94 pKa = 6.96PVAAIRR100 pKa = 11.84HH101 pKa = 5.39ALLTLRR107 pKa = 11.84EE108 pKa = 4.14KK109 pKa = 10.79KK110 pKa = 10.28RR111 pKa = 11.84LEE113 pKa = 4.04GEE115 pKa = 4.01RR116 pKa = 3.84
Molecular weight: 13.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
844879 |
33 |
3140 |
304.7 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.019 ± 0.061 | 1.763 ± 0.022 |
5.304 ± 0.035 | 6.968 ± 0.052 |
3.921 ± 0.033 | 8.524 ± 0.057 |
1.696 ± 0.02 | 5.272 ± 0.048 |
4.756 ± 0.051 | 10.37 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.025 | 2.878 ± 0.028 |
4.377 ± 0.036 | 3.729 ± 0.03 |
6.029 ± 0.044 | 5.369 ± 0.036 |
5.092 ± 0.033 | 7.128 ± 0.041 |
1.021 ± 0.018 | 3.388 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
