
Apis mellifera associated microvirus 33
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
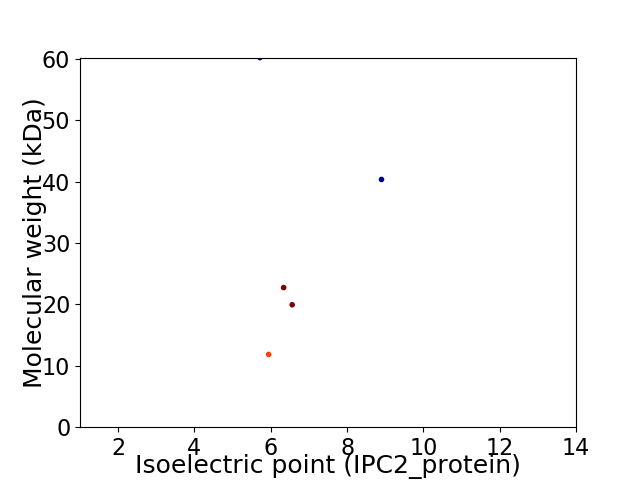
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTU5|A0A3S8UTU5_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 33 OX=2494762 PE=4 SV=1
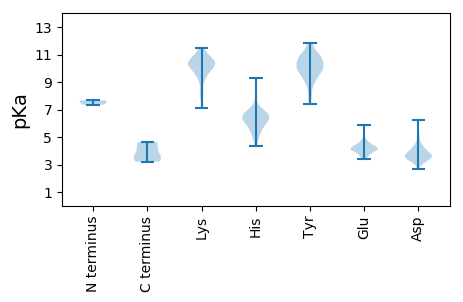
MM1 pKa = 7.35AHH3 pKa = 7.02LRR5 pKa = 11.84TDD7 pKa = 3.66TASNAHH13 pKa = 6.65FSRR16 pKa = 11.84VPTISRR22 pKa = 11.84SRR24 pKa = 11.84NAFSVAEE31 pKa = 4.04KK32 pKa = 10.51HH33 pKa = 4.9VTTSQFDD40 pKa = 3.77YY41 pKa = 10.84LYY43 pKa = 9.77PLYY46 pKa = 9.89WKK48 pKa = 10.23YY49 pKa = 10.7IYY51 pKa = 10.44PGDD54 pKa = 4.32TISLTHH60 pKa = 6.51GLLARR65 pKa = 11.84LQTQIAVLYY74 pKa = 10.28DD75 pKa = 3.59DD76 pKa = 5.99LYY78 pKa = 11.71VDD80 pKa = 3.31LHH82 pKa = 6.78AWFVPFRR89 pKa = 11.84LLQTNWARR97 pKa = 11.84YY98 pKa = 7.43QFNAQPLGPSQDD110 pKa = 3.49NSSLTSPKK118 pKa = 8.89ITLSGLGAGGFTAKK132 pKa = 10.21SLYY135 pKa = 10.39DD136 pKa = 3.43YY137 pKa = 10.99FGFPTEE143 pKa = 4.54INLSADD149 pKa = 3.49TQHH152 pKa = 6.45INNYY156 pKa = 9.69LGRR159 pKa = 11.84AYY161 pKa = 11.11NLIWNEE167 pKa = 3.67NYY169 pKa = 10.0RR170 pKa = 11.84DD171 pKa = 3.74QNLQNPATVDD181 pKa = 3.93LDD183 pKa = 4.42DD184 pKa = 6.26GPDD187 pKa = 3.4NPADD191 pKa = 3.69YY192 pKa = 10.74ILRR195 pKa = 11.84KK196 pKa = 9.23RR197 pKa = 11.84GKK199 pKa = 8.6RR200 pKa = 11.84HH201 pKa = 6.84DD202 pKa = 4.15RR203 pKa = 11.84FTSALTSQQKK213 pKa = 8.05GTAVTIPLGTSAPVLGIGTNTVQSWTAGPLTVNEE247 pKa = 4.35SDD249 pKa = 3.71LTTSQYY255 pKa = 11.86ANYY258 pKa = 10.35LLFRR262 pKa = 11.84GGSSDD267 pKa = 4.34IYY269 pKa = 11.01LEE271 pKa = 4.18KK272 pKa = 10.61QGSNNAPNIRR282 pKa = 11.84ADD284 pKa = 3.72LTGATAATLNQLRR297 pKa = 11.84QSVAVQHH304 pKa = 6.46LLEE307 pKa = 5.35ADD309 pKa = 3.31ARR311 pKa = 11.84GGTRR315 pKa = 11.84DD316 pKa = 3.03VEE318 pKa = 4.66AIHH321 pKa = 6.16NRR323 pKa = 11.84WGVVVPDD330 pKa = 4.74FRR332 pKa = 11.84LQRR335 pKa = 11.84PEE337 pKa = 3.83YY338 pKa = 10.63LGGQTFTFDD347 pKa = 3.29GHH349 pKa = 6.22VVPQTSATEE358 pKa = 4.1VGGTPQANLTQFSQAMTHH376 pKa = 6.27FNVNHH381 pKa = 6.28SFVEE385 pKa = 4.02HH386 pKa = 5.61GVYY389 pKa = 8.93MVLASARR396 pKa = 11.84SNITYY401 pKa = 9.62QQGLLRR407 pKa = 11.84EE408 pKa = 4.19LSYY411 pKa = 9.95RR412 pKa = 11.84TRR414 pKa = 11.84YY415 pKa = 10.11DD416 pKa = 2.65WFQPEE421 pKa = 4.13FANLGEE427 pKa = 4.09VAIKK431 pKa = 10.72NKK433 pKa = 9.53EE434 pKa = 3.62IYY436 pKa = 9.44MDD438 pKa = 4.3GSSADD443 pKa = 3.45EE444 pKa = 4.24STFGYY449 pKa = 9.96QEE451 pKa = 3.6YY452 pKa = 9.95AYY454 pKa = 9.72EE455 pKa = 4.46LRR457 pKa = 11.84YY458 pKa = 10.77SMNRR462 pKa = 11.84VTAEE466 pKa = 3.58MRR468 pKa = 11.84SNYY471 pKa = 8.05ATSLDD476 pKa = 4.02SKK478 pKa = 11.16HH479 pKa = 6.25MSDD482 pKa = 5.0EE483 pKa = 4.31YY484 pKa = 11.65ASLPTLGSAWIQSDD498 pKa = 3.97TPISRR503 pKa = 11.84NIAVSAATADD513 pKa = 4.25PIEE516 pKa = 4.74INTMAKK522 pKa = 10.37GMIARR527 pKa = 11.84TLPMYY532 pKa = 10.13SVPGLMRR539 pKa = 11.84LL540 pKa = 3.55
MM1 pKa = 7.35AHH3 pKa = 7.02LRR5 pKa = 11.84TDD7 pKa = 3.66TASNAHH13 pKa = 6.65FSRR16 pKa = 11.84VPTISRR22 pKa = 11.84SRR24 pKa = 11.84NAFSVAEE31 pKa = 4.04KK32 pKa = 10.51HH33 pKa = 4.9VTTSQFDD40 pKa = 3.77YY41 pKa = 10.84LYY43 pKa = 9.77PLYY46 pKa = 9.89WKK48 pKa = 10.23YY49 pKa = 10.7IYY51 pKa = 10.44PGDD54 pKa = 4.32TISLTHH60 pKa = 6.51GLLARR65 pKa = 11.84LQTQIAVLYY74 pKa = 10.28DD75 pKa = 3.59DD76 pKa = 5.99LYY78 pKa = 11.71VDD80 pKa = 3.31LHH82 pKa = 6.78AWFVPFRR89 pKa = 11.84LLQTNWARR97 pKa = 11.84YY98 pKa = 7.43QFNAQPLGPSQDD110 pKa = 3.49NSSLTSPKK118 pKa = 8.89ITLSGLGAGGFTAKK132 pKa = 10.21SLYY135 pKa = 10.39DD136 pKa = 3.43YY137 pKa = 10.99FGFPTEE143 pKa = 4.54INLSADD149 pKa = 3.49TQHH152 pKa = 6.45INNYY156 pKa = 9.69LGRR159 pKa = 11.84AYY161 pKa = 11.11NLIWNEE167 pKa = 3.67NYY169 pKa = 10.0RR170 pKa = 11.84DD171 pKa = 3.74QNLQNPATVDD181 pKa = 3.93LDD183 pKa = 4.42DD184 pKa = 6.26GPDD187 pKa = 3.4NPADD191 pKa = 3.69YY192 pKa = 10.74ILRR195 pKa = 11.84KK196 pKa = 9.23RR197 pKa = 11.84GKK199 pKa = 8.6RR200 pKa = 11.84HH201 pKa = 6.84DD202 pKa = 4.15RR203 pKa = 11.84FTSALTSQQKK213 pKa = 8.05GTAVTIPLGTSAPVLGIGTNTVQSWTAGPLTVNEE247 pKa = 4.35SDD249 pKa = 3.71LTTSQYY255 pKa = 11.86ANYY258 pKa = 10.35LLFRR262 pKa = 11.84GGSSDD267 pKa = 4.34IYY269 pKa = 11.01LEE271 pKa = 4.18KK272 pKa = 10.61QGSNNAPNIRR282 pKa = 11.84ADD284 pKa = 3.72LTGATAATLNQLRR297 pKa = 11.84QSVAVQHH304 pKa = 6.46LLEE307 pKa = 5.35ADD309 pKa = 3.31ARR311 pKa = 11.84GGTRR315 pKa = 11.84DD316 pKa = 3.03VEE318 pKa = 4.66AIHH321 pKa = 6.16NRR323 pKa = 11.84WGVVVPDD330 pKa = 4.74FRR332 pKa = 11.84LQRR335 pKa = 11.84PEE337 pKa = 3.83YY338 pKa = 10.63LGGQTFTFDD347 pKa = 3.29GHH349 pKa = 6.22VVPQTSATEE358 pKa = 4.1VGGTPQANLTQFSQAMTHH376 pKa = 6.27FNVNHH381 pKa = 6.28SFVEE385 pKa = 4.02HH386 pKa = 5.61GVYY389 pKa = 8.93MVLASARR396 pKa = 11.84SNITYY401 pKa = 9.62QQGLLRR407 pKa = 11.84EE408 pKa = 4.19LSYY411 pKa = 9.95RR412 pKa = 11.84TRR414 pKa = 11.84YY415 pKa = 10.11DD416 pKa = 2.65WFQPEE421 pKa = 4.13FANLGEE427 pKa = 4.09VAIKK431 pKa = 10.72NKK433 pKa = 9.53EE434 pKa = 3.62IYY436 pKa = 9.44MDD438 pKa = 4.3GSSADD443 pKa = 3.45EE444 pKa = 4.24STFGYY449 pKa = 9.96QEE451 pKa = 3.6YY452 pKa = 9.95AYY454 pKa = 9.72EE455 pKa = 4.46LRR457 pKa = 11.84YY458 pKa = 10.77SMNRR462 pKa = 11.84VTAEE466 pKa = 3.58MRR468 pKa = 11.84SNYY471 pKa = 8.05ATSLDD476 pKa = 4.02SKK478 pKa = 11.16HH479 pKa = 6.25MSDD482 pKa = 5.0EE483 pKa = 4.31YY484 pKa = 11.65ASLPTLGSAWIQSDD498 pKa = 3.97TPISRR503 pKa = 11.84NIAVSAATADD513 pKa = 4.25PIEE516 pKa = 4.74INTMAKK522 pKa = 10.37GMIARR527 pKa = 11.84TLPMYY532 pKa = 10.13SVPGLMRR539 pKa = 11.84LL540 pKa = 3.55
Molecular weight: 60.19 kDa
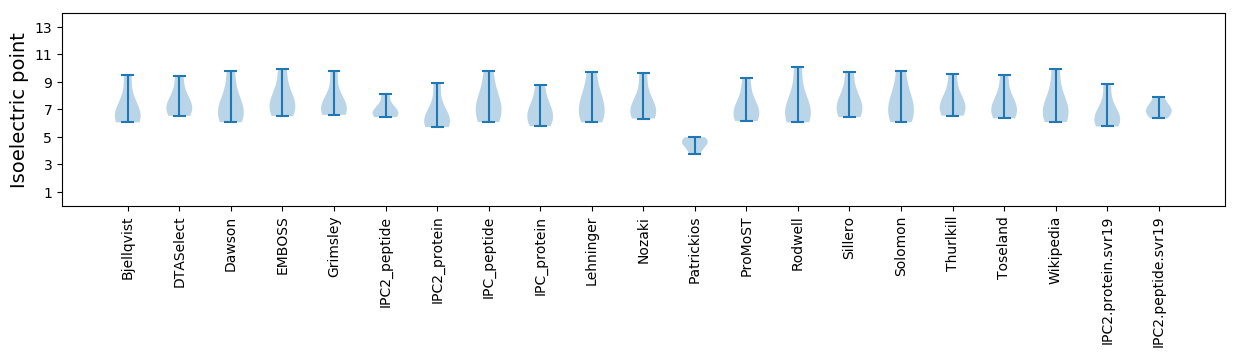
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTT5|A0A3S8UTT5_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 33 OX=2494762 PE=3 SV=1
MM1 pKa = 7.65EE2 pKa = 5.85KK3 pKa = 10.66GFSHH7 pKa = 6.61VKK9 pKa = 10.56CFDD12 pKa = 3.28PTLCYY17 pKa = 9.84TDD19 pKa = 3.96SRR21 pKa = 11.84GKK23 pKa = 10.01KK24 pKa = 9.14HH25 pKa = 5.66FRR27 pKa = 11.84HH28 pKa = 5.94YY29 pKa = 11.36SLANNLFLQMAQQVFDD45 pKa = 3.85CGKK48 pKa = 10.23CLRR51 pKa = 11.84CRR53 pKa = 11.84KK54 pKa = 9.83AKK56 pKa = 10.63ANEE59 pKa = 3.53LAARR63 pKa = 11.84CVLHH67 pKa = 7.88ASMYY71 pKa = 10.8DD72 pKa = 3.59SNCFLTLTYY81 pKa = 10.5DD82 pKa = 3.63EE83 pKa = 5.03KK84 pKa = 11.46KK85 pKa = 10.55EE86 pKa = 4.19GYY88 pKa = 9.68HH89 pKa = 6.89NNFNYY94 pKa = 10.63EE95 pKa = 4.03DD96 pKa = 3.53VQKK99 pKa = 10.78FKK101 pKa = 11.08KK102 pKa = 10.07RR103 pKa = 11.84LRR105 pKa = 11.84RR106 pKa = 11.84WVQYY110 pKa = 10.41HH111 pKa = 4.85FNKK114 pKa = 10.34RR115 pKa = 11.84IEE117 pKa = 4.02IFNVHH122 pKa = 6.48EE123 pKa = 4.0YY124 pKa = 10.84GKK126 pKa = 10.14NGKK129 pKa = 8.28KK130 pKa = 9.64HH131 pKa = 4.32YY132 pKa = 10.18HH133 pKa = 6.25LIVFNFDD140 pKa = 3.53FLDD143 pKa = 3.99KK144 pKa = 11.11EE145 pKa = 4.22IFTKK149 pKa = 10.87KK150 pKa = 10.36NGLPLYY156 pKa = 9.19TSKK159 pKa = 10.63ILEE162 pKa = 4.34KK163 pKa = 9.84RR164 pKa = 11.84WSHH167 pKa = 5.51GFCTIGDD174 pKa = 3.79VSEE177 pKa = 4.44ASAMYY182 pKa = 8.62QAKK185 pKa = 10.38YY186 pKa = 9.05MEE188 pKa = 5.28KK189 pKa = 10.25DD190 pKa = 3.55FQHH193 pKa = 6.88GNVTNSKK200 pKa = 9.84KK201 pKa = 10.33SHH203 pKa = 5.4SQHH206 pKa = 6.85SGIGRR211 pKa = 11.84PYY213 pKa = 8.95FLKK216 pKa = 10.47HH217 pKa = 5.3FRR219 pKa = 11.84QILSLGFIPFNGKK232 pKa = 9.43QIPVPRR238 pKa = 11.84YY239 pKa = 9.18FEE241 pKa = 4.26KK242 pKa = 10.63LAHH245 pKa = 6.62KK246 pKa = 9.92RR247 pKa = 11.84WCHH250 pKa = 5.53YY251 pKa = 10.06NDD253 pKa = 3.56PSAFVDD259 pKa = 4.52LPSRR263 pKa = 11.84DD264 pKa = 3.69ALYY267 pKa = 10.05TPFKK271 pKa = 10.01TGPLPYY277 pKa = 10.05SQLEE281 pKa = 3.88MSNLYY286 pKa = 9.3EE287 pKa = 4.1QYY289 pKa = 9.12KK290 pKa = 8.13TNKK293 pKa = 9.18DD294 pKa = 3.18AKK296 pKa = 8.19TQEE299 pKa = 4.62LAQEE303 pKa = 4.29WKK305 pKa = 10.95NVIQEE310 pKa = 4.09HH311 pKa = 6.84LKK313 pKa = 9.93TKK315 pKa = 10.22IPPDD319 pKa = 3.5FRR321 pKa = 11.84KK322 pKa = 10.4AGSNVLYY329 pKa = 10.43DD330 pKa = 3.84LKK332 pKa = 11.2NRR334 pKa = 11.84NNQEE338 pKa = 3.81KK339 pKa = 10.09FF340 pKa = 3.21
MM1 pKa = 7.65EE2 pKa = 5.85KK3 pKa = 10.66GFSHH7 pKa = 6.61VKK9 pKa = 10.56CFDD12 pKa = 3.28PTLCYY17 pKa = 9.84TDD19 pKa = 3.96SRR21 pKa = 11.84GKK23 pKa = 10.01KK24 pKa = 9.14HH25 pKa = 5.66FRR27 pKa = 11.84HH28 pKa = 5.94YY29 pKa = 11.36SLANNLFLQMAQQVFDD45 pKa = 3.85CGKK48 pKa = 10.23CLRR51 pKa = 11.84CRR53 pKa = 11.84KK54 pKa = 9.83AKK56 pKa = 10.63ANEE59 pKa = 3.53LAARR63 pKa = 11.84CVLHH67 pKa = 7.88ASMYY71 pKa = 10.8DD72 pKa = 3.59SNCFLTLTYY81 pKa = 10.5DD82 pKa = 3.63EE83 pKa = 5.03KK84 pKa = 11.46KK85 pKa = 10.55EE86 pKa = 4.19GYY88 pKa = 9.68HH89 pKa = 6.89NNFNYY94 pKa = 10.63EE95 pKa = 4.03DD96 pKa = 3.53VQKK99 pKa = 10.78FKK101 pKa = 11.08KK102 pKa = 10.07RR103 pKa = 11.84LRR105 pKa = 11.84RR106 pKa = 11.84WVQYY110 pKa = 10.41HH111 pKa = 4.85FNKK114 pKa = 10.34RR115 pKa = 11.84IEE117 pKa = 4.02IFNVHH122 pKa = 6.48EE123 pKa = 4.0YY124 pKa = 10.84GKK126 pKa = 10.14NGKK129 pKa = 8.28KK130 pKa = 9.64HH131 pKa = 4.32YY132 pKa = 10.18HH133 pKa = 6.25LIVFNFDD140 pKa = 3.53FLDD143 pKa = 3.99KK144 pKa = 11.11EE145 pKa = 4.22IFTKK149 pKa = 10.87KK150 pKa = 10.36NGLPLYY156 pKa = 9.19TSKK159 pKa = 10.63ILEE162 pKa = 4.34KK163 pKa = 9.84RR164 pKa = 11.84WSHH167 pKa = 5.51GFCTIGDD174 pKa = 3.79VSEE177 pKa = 4.44ASAMYY182 pKa = 8.62QAKK185 pKa = 10.38YY186 pKa = 9.05MEE188 pKa = 5.28KK189 pKa = 10.25DD190 pKa = 3.55FQHH193 pKa = 6.88GNVTNSKK200 pKa = 9.84KK201 pKa = 10.33SHH203 pKa = 5.4SQHH206 pKa = 6.85SGIGRR211 pKa = 11.84PYY213 pKa = 8.95FLKK216 pKa = 10.47HH217 pKa = 5.3FRR219 pKa = 11.84QILSLGFIPFNGKK232 pKa = 9.43QIPVPRR238 pKa = 11.84YY239 pKa = 9.18FEE241 pKa = 4.26KK242 pKa = 10.63LAHH245 pKa = 6.62KK246 pKa = 9.92RR247 pKa = 11.84WCHH250 pKa = 5.53YY251 pKa = 10.06NDD253 pKa = 3.56PSAFVDD259 pKa = 4.52LPSRR263 pKa = 11.84DD264 pKa = 3.69ALYY267 pKa = 10.05TPFKK271 pKa = 10.01TGPLPYY277 pKa = 10.05SQLEE281 pKa = 3.88MSNLYY286 pKa = 9.3EE287 pKa = 4.1QYY289 pKa = 9.12KK290 pKa = 8.13TNKK293 pKa = 9.18DD294 pKa = 3.18AKK296 pKa = 8.19TQEE299 pKa = 4.62LAQEE303 pKa = 4.29WKK305 pKa = 10.95NVIQEE310 pKa = 4.09HH311 pKa = 6.84LKK313 pKa = 9.93TKK315 pKa = 10.22IPPDD319 pKa = 3.5FRR321 pKa = 11.84KK322 pKa = 10.4AGSNVLYY329 pKa = 10.43DD330 pKa = 3.84LKK332 pKa = 11.2NRR334 pKa = 11.84NNQEE338 pKa = 3.81KK339 pKa = 10.09FF340 pKa = 3.21
Molecular weight: 40.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1370 |
104 |
540 |
274.0 |
31.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.394 ± 1.173 | 0.73 ± 0.587 |
5.109 ± 0.277 | 5.474 ± 1.087 |
4.599 ± 0.917 | 6.131 ± 0.888 |
3.139 ± 0.637 | 4.453 ± 0.551 |
6.715 ± 2.228 | 8.832 ± 0.346 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.847 ± 0.612 | 6.204 ± 0.38 |
4.234 ± 0.233 | 5.328 ± 0.221 |
4.891 ± 0.455 | 6.642 ± 1.261 |
6.423 ± 1.051 | 4.234 ± 0.428 |
0.876 ± 0.341 | 4.745 ± 0.729 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
