
Leishmania RNA virus 1 - 1 (isolate Leishmania guyanensis) (LRV-1-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Leishmaniavirus; Leishmania RNA virus 1
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
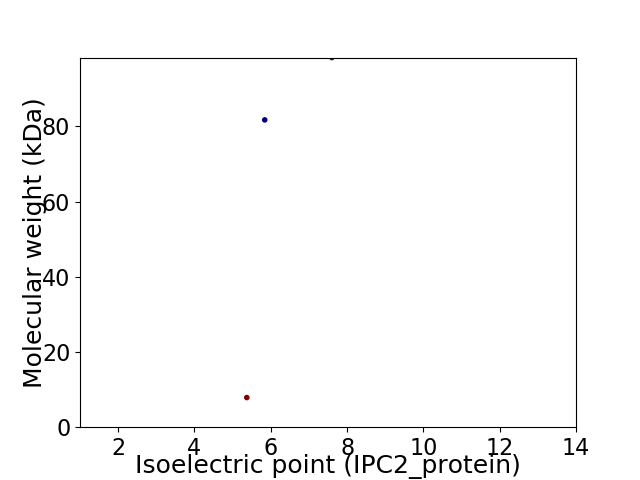
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q83096|ORF1_LRVLG Putative ORF1 protein OS=Leishmania RNA virus 1 - 1 (isolate Leishmania guyanensis) OX=58103 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.57PMSLDD6 pKa = 3.43SLAVGMVALLVDD18 pKa = 4.66YY19 pKa = 10.56YY20 pKa = 11.64SKK22 pKa = 11.47ALRR25 pKa = 11.84ASLNSTGIDD34 pKa = 4.12RR35 pKa = 11.84RR36 pKa = 11.84WGDD39 pKa = 3.23QWPSTVEE46 pKa = 3.51LTGRR50 pKa = 11.84GVIEE54 pKa = 4.09WEE56 pKa = 4.19SPTFCICSSSRR67 pKa = 11.84STAIVV72 pKa = 2.95
MM1 pKa = 7.57PMSLDD6 pKa = 3.43SLAVGMVALLVDD18 pKa = 4.66YY19 pKa = 10.56YY20 pKa = 11.64SKK22 pKa = 11.47ALRR25 pKa = 11.84ASLNSTGIDD34 pKa = 4.12RR35 pKa = 11.84RR36 pKa = 11.84WGDD39 pKa = 3.23QWPSTVEE46 pKa = 3.51LTGRR50 pKa = 11.84GVIEE54 pKa = 4.09WEE56 pKa = 4.19SPTFCICSSSRR67 pKa = 11.84STAIVV72 pKa = 2.95
Molecular weight: 7.88 kDa
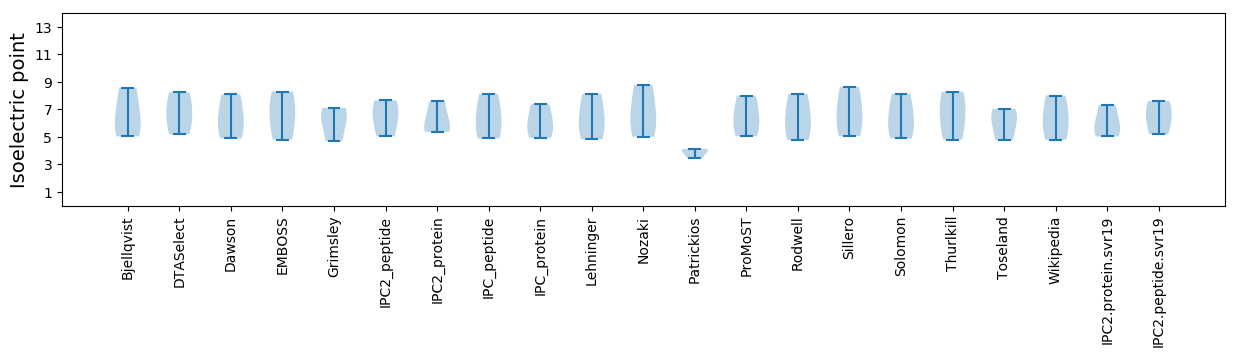
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q83096|ORF1_LRVLG Putative ORF1 protein OS=Leishmania RNA virus 1 - 1 (isolate Leishmania guyanensis) OX=58103 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.16QCPNQNHH8 pKa = 5.52MLVNRR13 pKa = 11.84AMVVAALDD21 pKa = 3.59SFEE24 pKa = 4.17DD25 pKa = 3.26ARR27 pKa = 11.84RR28 pKa = 11.84IISGVLDD35 pKa = 3.97LSRR38 pKa = 11.84LTNTSVKK45 pKa = 10.12GQHH48 pKa = 6.2TNDD51 pKa = 2.94TNYY54 pKa = 9.97FSRR57 pKa = 11.84YY58 pKa = 9.42SNFFSQMPAIILNQLKK74 pKa = 10.31CCIKK78 pKa = 10.69AQVDD82 pKa = 3.83DD83 pKa = 4.07AVKK86 pKa = 10.09MVLQKK91 pKa = 10.77AKK93 pKa = 10.03KK94 pKa = 9.2VEE96 pKa = 3.89VSKK99 pKa = 10.62PVEE102 pKa = 3.93KK103 pKa = 10.37MLTFTTLNTYY113 pKa = 10.4LGYY116 pKa = 9.74PEE118 pKa = 4.33STGCVMEE125 pKa = 4.15YY126 pKa = 10.15TEE128 pKa = 4.48EE129 pKa = 4.01QSGPIAAKK137 pKa = 10.65LLITLLSSTLNAMVRR152 pKa = 11.84PKK154 pKa = 10.46SDD156 pKa = 3.53PNISQNKK163 pKa = 8.27IPRR166 pKa = 11.84HH167 pKa = 4.79YY168 pKa = 9.8YY169 pKa = 7.43QLKK172 pKa = 8.43VHH174 pKa = 6.47VGAQKK179 pKa = 10.45KK180 pKa = 9.86VNLTAIEE187 pKa = 4.33VIRR190 pKa = 11.84GCQHH194 pKa = 5.66EE195 pKa = 4.59CSVVYY200 pKa = 10.58CEE202 pKa = 4.09FVRR205 pKa = 11.84YY206 pKa = 8.79SAYY209 pKa = 10.32FAGLYY214 pKa = 9.93DD215 pKa = 4.16DD216 pKa = 4.43QVAAILLYY224 pKa = 10.65AVAAHH229 pKa = 5.94NVQGFGARR237 pKa = 11.84FCVLWALMCVRR248 pKa = 11.84IAGFADD254 pKa = 5.0DD255 pKa = 3.31INIYY259 pKa = 9.79IKK261 pKa = 10.48HH262 pKa = 6.47RR263 pKa = 11.84GMSGLLPQLVEE274 pKa = 4.36MKK276 pKa = 10.18CLLGRR281 pKa = 11.84GVNEE285 pKa = 3.69IDD287 pKa = 3.62VEE289 pKa = 4.53TEE291 pKa = 3.06ARR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84LDD297 pKa = 3.31VGSLSMQRR305 pKa = 11.84LDD307 pKa = 3.83EE308 pKa = 4.48NEE310 pKa = 3.35LRR312 pKa = 11.84AAVRR316 pKa = 11.84LIYY319 pKa = 10.34SEE321 pKa = 4.04EE322 pKa = 3.73LRR324 pKa = 11.84RR325 pKa = 11.84PVTYY329 pKa = 10.15PLICDD334 pKa = 4.53FWSSRR339 pKa = 11.84WLWAANGSHH348 pKa = 6.18SRR350 pKa = 11.84ALEE353 pKa = 4.05HH354 pKa = 6.54AHH356 pKa = 7.1PEE358 pKa = 3.62LATRR362 pKa = 11.84KK363 pKa = 9.35EE364 pKa = 3.84GQAYY368 pKa = 9.18RR369 pKa = 11.84KK370 pKa = 10.0AVMEE374 pKa = 4.05QWQHH378 pKa = 6.38NPMDD382 pKa = 3.62RR383 pKa = 11.84WDD385 pKa = 3.51GTVYY389 pKa = 8.71VTPSAKK395 pKa = 10.15LEE397 pKa = 4.2HH398 pKa = 6.6GKK400 pKa = 7.98TRR402 pKa = 11.84LLLACDD408 pKa = 3.7TLSYY412 pKa = 10.0MWFEE416 pKa = 3.92YY417 pKa = 10.81ALRR420 pKa = 11.84PVEE423 pKa = 4.94RR424 pKa = 11.84IWEE427 pKa = 4.02NSNVILDD434 pKa = 4.11PGSMGNCGIATRR446 pKa = 11.84INGWRR451 pKa = 11.84NGMPGQSFFAVDD463 pKa = 3.44YY464 pKa = 11.42DD465 pKa = 4.57DD466 pKa = 6.21FNSQHH471 pKa = 5.85TLMSQKK477 pKa = 10.04IVFEE481 pKa = 4.21EE482 pKa = 4.33LFHH485 pKa = 7.46HH486 pKa = 6.79IGYY489 pKa = 8.57NASWVKK495 pKa = 9.77TLVDD499 pKa = 3.99SFDD502 pKa = 5.35SMEE505 pKa = 3.9LWIKK509 pKa = 9.08GKK511 pKa = 10.22CAGIMAGTLMSGHH524 pKa = 7.1RR525 pKa = 11.84ATSFINSVLNRR536 pKa = 11.84AYY538 pKa = 9.67IICAGGHH545 pKa = 5.89VPTSMHH551 pKa = 6.21VGDD554 pKa = 6.22DD555 pKa = 3.1ILMSCTLGHH564 pKa = 7.38ADD566 pKa = 4.54NLIANLNRR574 pKa = 11.84KK575 pKa = 7.49GVRR578 pKa = 11.84LNASKK583 pKa = 10.6QVFSKK588 pKa = 10.12TSGEE592 pKa = 4.28FLRR595 pKa = 11.84VAHH598 pKa = 7.0RR599 pKa = 11.84EE600 pKa = 4.36HH601 pKa = 6.78TSHH604 pKa = 6.95GYY606 pKa = 8.27LARR609 pKa = 11.84VISSAVSGNWVSDD622 pKa = 3.2HH623 pKa = 6.44TLNQQEE629 pKa = 4.57ALMNAIVCCRR639 pKa = 11.84GILNRR644 pKa = 11.84SLPGEE649 pKa = 4.16KK650 pKa = 10.33NPVVRR655 pKa = 11.84VISRR659 pKa = 11.84SVSKK663 pKa = 9.68RR664 pKa = 11.84TKK666 pKa = 10.09IEE668 pKa = 3.79EE669 pKa = 3.82KK670 pKa = 10.11TIRR673 pKa = 11.84LLLSGRR679 pKa = 11.84ACLKK683 pKa = 10.22GGVVYY688 pKa = 10.74GEE690 pKa = 3.88QTNYY694 pKa = 8.69IQVYY698 pKa = 9.85RR699 pKa = 11.84INCRR703 pKa = 11.84VEE705 pKa = 3.88RR706 pKa = 11.84SEE708 pKa = 4.78EE709 pKa = 3.68KK710 pKa = 10.65LPPYY714 pKa = 10.25RR715 pKa = 11.84HH716 pKa = 5.69ATEE719 pKa = 6.02DD720 pKa = 3.53YY721 pKa = 10.92LNNHH725 pKa = 6.63LADD728 pKa = 3.64IEE730 pKa = 4.55VMAVRR735 pKa = 11.84QYY737 pKa = 11.7GSDD740 pKa = 3.01IADD743 pKa = 3.14IMAQASWKK751 pKa = 10.46KK752 pKa = 10.93SMSTEE757 pKa = 3.67GAEE760 pKa = 4.21DD761 pKa = 3.63VSRR764 pKa = 11.84LSLQRR769 pKa = 11.84DD770 pKa = 3.19KK771 pKa = 11.05TLPCLHH777 pKa = 7.23CITEE781 pKa = 4.54KK782 pKa = 9.9EE783 pKa = 4.4TSLLPVRR790 pKa = 11.84YY791 pKa = 9.16GLFSSYY797 pKa = 10.12PILMMLKK804 pKa = 9.99DD805 pKa = 4.93RR806 pKa = 11.84IPIKK810 pKa = 10.33EE811 pKa = 4.11ALKK814 pKa = 10.12LAVTIGYY821 pKa = 9.2RR822 pKa = 11.84PQPNSDD828 pKa = 4.25LEE830 pKa = 5.0LDD832 pKa = 3.65LWGEE836 pKa = 4.56SNNSCAIEE844 pKa = 4.4GVLPYY849 pKa = 10.92NEE851 pKa = 4.23ATSLAQKK858 pKa = 9.53LPCGGVVIQVIHH870 pKa = 5.75NVYY873 pKa = 9.98VV874 pKa = 3.24
MM1 pKa = 7.16QCPNQNHH8 pKa = 5.52MLVNRR13 pKa = 11.84AMVVAALDD21 pKa = 3.59SFEE24 pKa = 4.17DD25 pKa = 3.26ARR27 pKa = 11.84RR28 pKa = 11.84IISGVLDD35 pKa = 3.97LSRR38 pKa = 11.84LTNTSVKK45 pKa = 10.12GQHH48 pKa = 6.2TNDD51 pKa = 2.94TNYY54 pKa = 9.97FSRR57 pKa = 11.84YY58 pKa = 9.42SNFFSQMPAIILNQLKK74 pKa = 10.31CCIKK78 pKa = 10.69AQVDD82 pKa = 3.83DD83 pKa = 4.07AVKK86 pKa = 10.09MVLQKK91 pKa = 10.77AKK93 pKa = 10.03KK94 pKa = 9.2VEE96 pKa = 3.89VSKK99 pKa = 10.62PVEE102 pKa = 3.93KK103 pKa = 10.37MLTFTTLNTYY113 pKa = 10.4LGYY116 pKa = 9.74PEE118 pKa = 4.33STGCVMEE125 pKa = 4.15YY126 pKa = 10.15TEE128 pKa = 4.48EE129 pKa = 4.01QSGPIAAKK137 pKa = 10.65LLITLLSSTLNAMVRR152 pKa = 11.84PKK154 pKa = 10.46SDD156 pKa = 3.53PNISQNKK163 pKa = 8.27IPRR166 pKa = 11.84HH167 pKa = 4.79YY168 pKa = 9.8YY169 pKa = 7.43QLKK172 pKa = 8.43VHH174 pKa = 6.47VGAQKK179 pKa = 10.45KK180 pKa = 9.86VNLTAIEE187 pKa = 4.33VIRR190 pKa = 11.84GCQHH194 pKa = 5.66EE195 pKa = 4.59CSVVYY200 pKa = 10.58CEE202 pKa = 4.09FVRR205 pKa = 11.84YY206 pKa = 8.79SAYY209 pKa = 10.32FAGLYY214 pKa = 9.93DD215 pKa = 4.16DD216 pKa = 4.43QVAAILLYY224 pKa = 10.65AVAAHH229 pKa = 5.94NVQGFGARR237 pKa = 11.84FCVLWALMCVRR248 pKa = 11.84IAGFADD254 pKa = 5.0DD255 pKa = 3.31INIYY259 pKa = 9.79IKK261 pKa = 10.48HH262 pKa = 6.47RR263 pKa = 11.84GMSGLLPQLVEE274 pKa = 4.36MKK276 pKa = 10.18CLLGRR281 pKa = 11.84GVNEE285 pKa = 3.69IDD287 pKa = 3.62VEE289 pKa = 4.53TEE291 pKa = 3.06ARR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84LDD297 pKa = 3.31VGSLSMQRR305 pKa = 11.84LDD307 pKa = 3.83EE308 pKa = 4.48NEE310 pKa = 3.35LRR312 pKa = 11.84AAVRR316 pKa = 11.84LIYY319 pKa = 10.34SEE321 pKa = 4.04EE322 pKa = 3.73LRR324 pKa = 11.84RR325 pKa = 11.84PVTYY329 pKa = 10.15PLICDD334 pKa = 4.53FWSSRR339 pKa = 11.84WLWAANGSHH348 pKa = 6.18SRR350 pKa = 11.84ALEE353 pKa = 4.05HH354 pKa = 6.54AHH356 pKa = 7.1PEE358 pKa = 3.62LATRR362 pKa = 11.84KK363 pKa = 9.35EE364 pKa = 3.84GQAYY368 pKa = 9.18RR369 pKa = 11.84KK370 pKa = 10.0AVMEE374 pKa = 4.05QWQHH378 pKa = 6.38NPMDD382 pKa = 3.62RR383 pKa = 11.84WDD385 pKa = 3.51GTVYY389 pKa = 8.71VTPSAKK395 pKa = 10.15LEE397 pKa = 4.2HH398 pKa = 6.6GKK400 pKa = 7.98TRR402 pKa = 11.84LLLACDD408 pKa = 3.7TLSYY412 pKa = 10.0MWFEE416 pKa = 3.92YY417 pKa = 10.81ALRR420 pKa = 11.84PVEE423 pKa = 4.94RR424 pKa = 11.84IWEE427 pKa = 4.02NSNVILDD434 pKa = 4.11PGSMGNCGIATRR446 pKa = 11.84INGWRR451 pKa = 11.84NGMPGQSFFAVDD463 pKa = 3.44YY464 pKa = 11.42DD465 pKa = 4.57DD466 pKa = 6.21FNSQHH471 pKa = 5.85TLMSQKK477 pKa = 10.04IVFEE481 pKa = 4.21EE482 pKa = 4.33LFHH485 pKa = 7.46HH486 pKa = 6.79IGYY489 pKa = 8.57NASWVKK495 pKa = 9.77TLVDD499 pKa = 3.99SFDD502 pKa = 5.35SMEE505 pKa = 3.9LWIKK509 pKa = 9.08GKK511 pKa = 10.22CAGIMAGTLMSGHH524 pKa = 7.1RR525 pKa = 11.84ATSFINSVLNRR536 pKa = 11.84AYY538 pKa = 9.67IICAGGHH545 pKa = 5.89VPTSMHH551 pKa = 6.21VGDD554 pKa = 6.22DD555 pKa = 3.1ILMSCTLGHH564 pKa = 7.38ADD566 pKa = 4.54NLIANLNRR574 pKa = 11.84KK575 pKa = 7.49GVRR578 pKa = 11.84LNASKK583 pKa = 10.6QVFSKK588 pKa = 10.12TSGEE592 pKa = 4.28FLRR595 pKa = 11.84VAHH598 pKa = 7.0RR599 pKa = 11.84EE600 pKa = 4.36HH601 pKa = 6.78TSHH604 pKa = 6.95GYY606 pKa = 8.27LARR609 pKa = 11.84VISSAVSGNWVSDD622 pKa = 3.2HH623 pKa = 6.44TLNQQEE629 pKa = 4.57ALMNAIVCCRR639 pKa = 11.84GILNRR644 pKa = 11.84SLPGEE649 pKa = 4.16KK650 pKa = 10.33NPVVRR655 pKa = 11.84VISRR659 pKa = 11.84SVSKK663 pKa = 9.68RR664 pKa = 11.84TKK666 pKa = 10.09IEE668 pKa = 3.79EE669 pKa = 3.82KK670 pKa = 10.11TIRR673 pKa = 11.84LLLSGRR679 pKa = 11.84ACLKK683 pKa = 10.22GGVVYY688 pKa = 10.74GEE690 pKa = 3.88QTNYY694 pKa = 8.69IQVYY698 pKa = 9.85RR699 pKa = 11.84INCRR703 pKa = 11.84VEE705 pKa = 3.88RR706 pKa = 11.84SEE708 pKa = 4.78EE709 pKa = 3.68KK710 pKa = 10.65LPPYY714 pKa = 10.25RR715 pKa = 11.84HH716 pKa = 5.69ATEE719 pKa = 6.02DD720 pKa = 3.53YY721 pKa = 10.92LNNHH725 pKa = 6.63LADD728 pKa = 3.64IEE730 pKa = 4.55VMAVRR735 pKa = 11.84QYY737 pKa = 11.7GSDD740 pKa = 3.01IADD743 pKa = 3.14IMAQASWKK751 pKa = 10.46KK752 pKa = 10.93SMSTEE757 pKa = 3.67GAEE760 pKa = 4.21DD761 pKa = 3.63VSRR764 pKa = 11.84LSLQRR769 pKa = 11.84DD770 pKa = 3.19KK771 pKa = 11.05TLPCLHH777 pKa = 7.23CITEE781 pKa = 4.54KK782 pKa = 9.9EE783 pKa = 4.4TSLLPVRR790 pKa = 11.84YY791 pKa = 9.16GLFSSYY797 pKa = 10.12PILMMLKK804 pKa = 9.99DD805 pKa = 4.93RR806 pKa = 11.84IPIKK810 pKa = 10.33EE811 pKa = 4.11ALKK814 pKa = 10.12LAVTIGYY821 pKa = 9.2RR822 pKa = 11.84PQPNSDD828 pKa = 4.25LEE830 pKa = 5.0LDD832 pKa = 3.65LWGEE836 pKa = 4.56SNNSCAIEE844 pKa = 4.4GVLPYY849 pKa = 10.92NEE851 pKa = 4.23ATSLAQKK858 pKa = 9.53LPCGGVVIQVIHH870 pKa = 5.75NVYY873 pKa = 9.98VV874 pKa = 3.24
Molecular weight: 98.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1687 |
72 |
874 |
562.3 |
62.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.773 ± 1.014 | 2.845 ± 0.078 |
5.098 ± 0.65 | 4.861 ± 0.646 |
2.845 ± 0.359 | 6.106 ± 0.125 |
3.023 ± 0.45 | 5.513 ± 0.247 |
3.794 ± 0.794 | 9.247 ± 0.469 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.142 ± 0.221 | 4.92 ± 0.552 |
4.149 ± 0.545 | 3.142 ± 0.51 |
6.165 ± 0.165 | 7.647 ± 1.139 |
5.868 ± 0.97 | 7.291 ± 0.51 |
1.541 ± 0.406 | 4.031 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
