
Sulfuricurvum kujiense (strain ATCC BAA-921 / DSM 16994 / JCM 11577 / YK-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Thiovulaceae; Sulfuricurvum; Sulfuricurvum kujiense
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
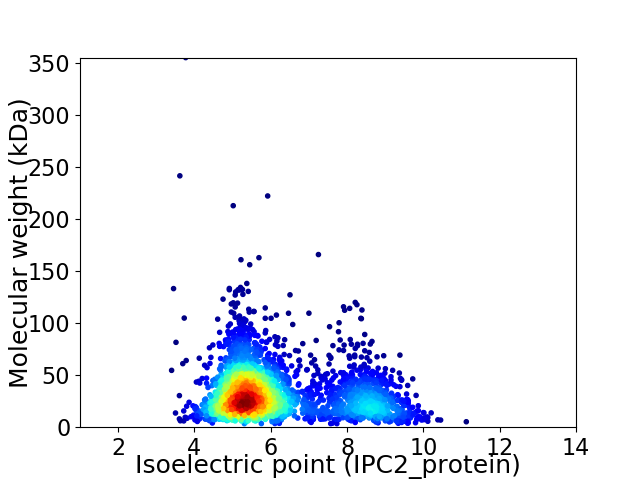
Virtual 2D-PAGE plot for 2794 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4U0I0|E4U0I0_SULKY Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor OS=Sulfuricurvum kujiense (strain ATCC BAA-921 / DSM 16994 / JCM 11577 / YK-1) OX=709032 GN=Sulku_1636 PE=4 SV=1
MM1 pKa = 7.65DD2 pKa = 4.43TATPQQIAFILDD14 pKa = 3.33NVTDD18 pKa = 3.93YY19 pKa = 11.45QSLVEE24 pKa = 4.96GIPEE28 pKa = 4.17GTPVYY33 pKa = 10.66VLNNVDD39 pKa = 5.18DD40 pKa = 4.44VLAQIAAITADD51 pKa = 3.92YY52 pKa = 11.21SNLDD56 pKa = 4.17AIHH59 pKa = 6.58LLSHH63 pKa = 6.74GSDD66 pKa = 3.36GSINLGTFSLNSDD79 pKa = 4.0NLNTYY84 pKa = 11.04ADD86 pKa = 3.69TLGQIGASLTEE97 pKa = 4.43HH98 pKa = 7.14GDD100 pKa = 3.16ILLYY104 pKa = 10.03GCNVARR110 pKa = 11.84DD111 pKa = 3.45QSGIDD116 pKa = 3.76FIGKK120 pKa = 8.99LAQLSGADD128 pKa = 3.14IAASNDD134 pKa = 3.26MTGSNTQNGDD144 pKa = 3.58WILEE148 pKa = 4.12SSIGMIDD155 pKa = 3.51TPALVLNQYY164 pKa = 9.56NQTLAAGTVTFTSSPSAATATDD186 pKa = 3.73GTASNDD192 pKa = 2.91IAGIVLNFTSDD203 pKa = 3.94LGTNWTYY210 pKa = 7.38EE211 pKa = 4.03TPYY214 pKa = 8.64TGSVITVGYY223 pKa = 7.85ATNAPQALMTIKK235 pKa = 10.7SSDD238 pKa = 3.19GSNFWFKK245 pKa = 11.42SLFIADD251 pKa = 3.83SGGQPVKK258 pKa = 11.24VEE260 pKa = 3.96GFEE263 pKa = 4.68NNVSTGSVNLDD274 pKa = 3.33TTGGGSLYY282 pKa = 10.67EE283 pKa = 4.19NTFGTASLTQSIFQNVDD300 pKa = 2.86EE301 pKa = 4.86VRR303 pKa = 11.84ITPQSGVQMWIGVNNIQIDD322 pKa = 4.12NPVLPGPTVTSALYY336 pKa = 10.22DD337 pKa = 3.4ASTNSLVVTGTNMTATAGVTNDD359 pKa = 2.9INVAKK364 pKa = 9.37LTLTGEE370 pKa = 4.37GGATYY375 pKa = 9.95TLTSSNVEE383 pKa = 3.73IDD385 pKa = 3.2SATQFTVALNATDD398 pKa = 3.69QINVEE403 pKa = 4.0GLLNKK408 pKa = 10.43NGTSSVGTTTYY419 pKa = 10.9NIAAAADD426 pKa = 3.5WDD428 pKa = 4.19VASTGNADD436 pKa = 3.44LTGNGITVSNTQTPTITSATYY457 pKa = 10.28DD458 pKa = 3.45AATGSLVVTGTNLVKK473 pKa = 8.89TTGATNDD480 pKa = 2.94ITVNKK485 pKa = 8.93LTFTGEE491 pKa = 4.0GGSTYY496 pKa = 10.6TLTSSNVEE504 pKa = 3.77ITDD507 pKa = 3.37ATSFSVTLNATDD519 pKa = 4.66LAALNQIVNKK529 pKa = 10.28NGTSSTGATTYY540 pKa = 11.38NLAAADD546 pKa = 4.09DD547 pKa = 4.34WNSVITAGTIADD559 pKa = 3.6ATSAVTASNVAVPAITNATYY579 pKa = 10.47DD580 pKa = 3.41ASTGALVVTGTGFLKK595 pKa = 10.96LSGATNDD602 pKa = 2.83IVANKK607 pKa = 8.26FTFTGEE613 pKa = 3.66GGATYY618 pKa = 10.2TLTDD622 pKa = 3.52TSNVEE627 pKa = 4.09ITSGTAFTITLSATDD642 pKa = 3.78KK643 pKa = 11.15AALNLMMDD651 pKa = 3.84KK652 pKa = 10.92NGTSSTSGTTYY663 pKa = 11.3NLAAAEE669 pKa = 4.06DD670 pKa = 3.91WAAGADD676 pKa = 3.67AAVVVADD683 pKa = 3.76TTGNGVTASNVAVPTITSSTYY704 pKa = 9.72NASTGALVVTGTGFLSKK721 pKa = 10.63SGATNDD727 pKa = 2.69IVANKK732 pKa = 8.26FTFTGEE738 pKa = 3.66GGATYY743 pKa = 10.2TLTDD747 pKa = 3.52TSNVEE752 pKa = 3.93ITSGTVFTITLSATDD767 pKa = 4.1LAGVDD772 pKa = 3.95QIMNKK777 pKa = 10.09NGTASTSGTTYY788 pKa = 11.3NLAAAEE794 pKa = 4.27DD795 pKa = 4.02WASGADD801 pKa = 3.49AAVVVADD808 pKa = 3.76TTGNGVTASNVAVPTITSATYY829 pKa = 10.15DD830 pKa = 3.54YY831 pKa = 8.69NTNVLTVTGTGFLNKK846 pKa = 10.07SGATNDD852 pKa = 2.97IDD854 pKa = 3.98LTKK857 pKa = 10.2LTFSGEE863 pKa = 4.13GGSTYY868 pKa = 10.77TLTNATGVEE877 pKa = 4.3ITSGTSFSVTLSGADD892 pKa = 3.96LINVEE897 pKa = 4.3TLLNKK902 pKa = 10.41NGTSAVSGTTYY913 pKa = 11.3NLAAAEE919 pKa = 4.06DD920 pKa = 3.91WAAGADD926 pKa = 3.67AAVVVADD933 pKa = 3.85TTGNGITLSNYY944 pKa = 8.44AAPTVTSATYY954 pKa = 10.34DD955 pKa = 3.04AGTHH959 pKa = 4.84VLSVTGTNLVSKK971 pKa = 10.72SGATNDD977 pKa = 3.49VAVSMLTVTGEE988 pKa = 3.96GGTYY992 pKa = 9.67TLTSSDD998 pKa = 3.36VDD1000 pKa = 3.25ITNATTFSVTLNAADD1015 pKa = 3.46QLVVNGLLNKK1025 pKa = 10.44NGTASSGATTYY1036 pKa = 11.46NLAAAEE1042 pKa = 4.14DD1043 pKa = 4.21WLAGTASATVVADD1056 pKa = 3.44LTGNGITVSNVQTPTITSATYY1077 pKa = 10.77DD1078 pKa = 3.64STTGSLVVTGTNFFSKK1094 pKa = 10.58VGANNDD1100 pKa = 2.86IDD1102 pKa = 4.22LTKK1105 pKa = 10.39LTFTGGAANATYY1117 pKa = 9.71TLSSTSNVEE1126 pKa = 3.67ITSATSFSITLSGADD1141 pKa = 3.58KK1142 pKa = 10.71TSVDD1146 pKa = 3.97ALLDD1150 pKa = 4.3NIGTTSSGGSTYY1162 pKa = 11.36NLAAANNWLAGADD1175 pKa = 3.61SAAVITDD1182 pKa = 3.74AVNAITVSINPAITSATYY1200 pKa = 10.2DD1201 pKa = 3.23ASTGSLVVTGTNMQVMAGAANDD1223 pKa = 3.19ITANKK1228 pKa = 8.7LTFTGEE1234 pKa = 3.65GGATYY1239 pKa = 10.15TLTDD1243 pKa = 3.5SANVEE1248 pKa = 4.17LSSATSFTIALSATDD1263 pKa = 3.83KK1264 pKa = 11.25AAINQIVNKK1273 pKa = 9.96NGTSSTGATTYY1284 pKa = 11.38NLAAADD1290 pKa = 3.73DD1291 pKa = 4.16WNAQVSAGDD1300 pKa = 3.57TSDD1303 pKa = 2.98ATNAVTVSSVAAPTITNATYY1323 pKa = 10.44DD1324 pKa = 3.59ASTGALVVTGTGFLKK1339 pKa = 10.96LSGATNDD1346 pKa = 2.83IVANKK1351 pKa = 8.26FTFTGEE1357 pKa = 3.66GGATYY1362 pKa = 10.2TLTDD1366 pKa = 3.52TSNVEE1371 pKa = 4.09ITSGTAFTITLSATDD1386 pKa = 3.78KK1387 pKa = 11.15AALNLMMDD1395 pKa = 3.84KK1396 pKa = 10.92NGTSSTSGTTYY1407 pKa = 11.3NLAAAEE1413 pKa = 4.06DD1414 pKa = 3.91WAAGADD1420 pKa = 3.67AAVVVADD1427 pKa = 3.76TTGNGVTASNVAVPTITSSTYY1448 pKa = 9.72NASTGALVVTGTGFLSKK1465 pKa = 10.63SGATNDD1471 pKa = 2.69IVANKK1476 pKa = 8.26FTFTGEE1482 pKa = 3.66GGATYY1487 pKa = 10.2TLTDD1491 pKa = 3.52TSNVEE1496 pKa = 3.93ITSGTVFTITLSATDD1511 pKa = 4.1LAGVDD1516 pKa = 3.95QIMNKK1521 pKa = 10.09NGTASTSGTTYY1532 pKa = 11.3NLAAAEE1538 pKa = 4.27DD1539 pKa = 4.02WASGADD1545 pKa = 3.49AAVVVADD1552 pKa = 3.76TTGNGVTASNVAVPAITSSTYY1573 pKa = 10.09NASTGALVVTGTGFLKK1589 pKa = 10.96LSGATNDD1596 pKa = 2.83IVANKK1601 pKa = 8.26FTFTGEE1607 pKa = 3.66GGATYY1612 pKa = 10.2TLTDD1616 pKa = 3.52TSNVEE1621 pKa = 4.09ITSGTAFTITLSATDD1636 pKa = 3.78KK1637 pKa = 11.15AALNLMMDD1645 pKa = 3.84KK1646 pKa = 10.92NGTSSTSGTTYY1657 pKa = 11.3NLAAAEE1663 pKa = 4.06DD1664 pKa = 3.91WAAGADD1670 pKa = 3.67AAVVVADD1677 pKa = 3.76TTGNGVTASNVAVPTITSSTYY1698 pKa = 9.72NASTGALVVTGTGFLSKK1715 pKa = 10.63SGATNDD1721 pKa = 2.69IVANKK1726 pKa = 8.26FTFTGEE1732 pKa = 3.66GGATYY1737 pKa = 10.2TLTDD1741 pKa = 3.52TSNVEE1746 pKa = 3.93ITSGTVFTITLSATDD1761 pKa = 4.1LAGVDD1766 pKa = 3.95QIMNKK1771 pKa = 10.09NGTASTSGTTYY1782 pKa = 11.3NLAAAEE1788 pKa = 4.27DD1789 pKa = 4.02WASGADD1795 pKa = 3.49AAVVVADD1802 pKa = 3.76TTGNGVTASNVAVPTITSATYY1823 pKa = 10.5DD1824 pKa = 3.25SGTGVLTVSGTHH1836 pKa = 5.88FVHH1839 pKa = 7.59KK1840 pKa = 10.46SGSTNDD1846 pKa = 3.44IDD1848 pKa = 6.19LSMLTFTGEE1857 pKa = 3.7AGGTYY1862 pKa = 9.82TLTTATDD1869 pKa = 3.53VEE1871 pKa = 4.73ITSGTSFTVTLSGADD1886 pKa = 3.31KK1887 pKa = 10.74TGVDD1891 pKa = 3.42TLLNKK1896 pKa = 10.11IGTSSTDD1903 pKa = 2.72ATTYY1907 pKa = 11.2NLAAAEE1913 pKa = 4.12DD1914 pKa = 4.16WATGADD1920 pKa = 3.26AAVNVVDD1927 pKa = 3.91ATNAITVSQTIASSGGGGGTSTPTTTIDD1955 pKa = 3.24GATVEE1960 pKa = 4.62TTTATNSSGQTVEE1973 pKa = 4.32QIIILPISSTRR1984 pKa = 11.84VNSTGDD1990 pKa = 3.05TATADD1995 pKa = 3.22IPLFWGEE2002 pKa = 3.87SSRR2005 pKa = 11.84TVDD2008 pKa = 2.86ATTANLPTGIGLSATGARR2026 pKa = 11.84APQSGHH2032 pKa = 6.45TIQDD2036 pKa = 3.83SINDD2040 pKa = 3.94LLALINEE2047 pKa = 4.78TAPSTDD2053 pKa = 3.04TSKK2056 pKa = 10.17TSMLGGGSTFLNTLSGITDD2075 pKa = 3.25NLVINKK2081 pKa = 7.94ITLTHH2086 pKa = 6.96TGTSAPGMPITIQGTTSTVTTSGGALPPVEE2116 pKa = 4.76ALVIDD2121 pKa = 4.47GQTLLSNTTLSLNNVEE2137 pKa = 4.28FATIIGEE2144 pKa = 4.0NLIIRR2149 pKa = 11.84GGEE2152 pKa = 3.79GRR2154 pKa = 11.84NTIFTGDD2161 pKa = 3.24GHH2163 pKa = 6.8QDD2165 pKa = 3.12IMCGTDD2171 pKa = 3.85DD2172 pKa = 5.74DD2173 pKa = 4.51EE2174 pKa = 4.61LHH2176 pKa = 7.08AGGGDD2181 pKa = 3.67DD2182 pKa = 3.82TVGSAGGNDD2191 pKa = 3.83RR2192 pKa = 11.84IFAEE2196 pKa = 4.64AGNDD2200 pKa = 3.54TVFGGTGDD2208 pKa = 4.27DD2209 pKa = 4.03FMHH2212 pKa = 6.75GGSGNDD2218 pKa = 2.96TVTYY2222 pKa = 10.2DD2223 pKa = 3.79GNRR2226 pKa = 11.84GDD2228 pKa = 4.28YY2229 pKa = 10.94VITRR2233 pKa = 11.84DD2234 pKa = 3.24NGKK2237 pKa = 7.74TYY2239 pKa = 10.75VYY2241 pKa = 9.47TVSTGEE2247 pKa = 3.8TDD2249 pKa = 3.49TVVNAEE2255 pKa = 4.49SIQFADD2261 pKa = 4.91AIYY2264 pKa = 9.56TITNNEE2270 pKa = 4.71DD2271 pKa = 3.02LTQIASLYY2279 pKa = 8.19QQILGRR2285 pKa = 11.84QADD2288 pKa = 4.02LDD2290 pKa = 3.88GFQYY2294 pKa = 7.64WTSTFDD2300 pKa = 3.27NGDD2303 pKa = 3.75TIGAIATSFVRR2314 pKa = 11.84SSEE2317 pKa = 4.06YY2318 pKa = 10.85FNDD2321 pKa = 3.7TGNVWDD2327 pKa = 5.19SMSNDD2332 pKa = 3.01QRR2334 pKa = 11.84IEE2336 pKa = 3.83TFYY2339 pKa = 11.42QLMLGRR2345 pKa = 11.84ASDD2348 pKa = 4.34AVGKK2352 pKa = 9.94AYY2354 pKa = 9.76WMDD2357 pKa = 5.06AINNGTMSIQDD2368 pKa = 3.25IAGSFVEE2375 pKa = 4.24SSEE2378 pKa = 3.83MQGIYY2383 pKa = 9.62TGQQAWSFFVV2393 pKa = 3.18
MM1 pKa = 7.65DD2 pKa = 4.43TATPQQIAFILDD14 pKa = 3.33NVTDD18 pKa = 3.93YY19 pKa = 11.45QSLVEE24 pKa = 4.96GIPEE28 pKa = 4.17GTPVYY33 pKa = 10.66VLNNVDD39 pKa = 5.18DD40 pKa = 4.44VLAQIAAITADD51 pKa = 3.92YY52 pKa = 11.21SNLDD56 pKa = 4.17AIHH59 pKa = 6.58LLSHH63 pKa = 6.74GSDD66 pKa = 3.36GSINLGTFSLNSDD79 pKa = 4.0NLNTYY84 pKa = 11.04ADD86 pKa = 3.69TLGQIGASLTEE97 pKa = 4.43HH98 pKa = 7.14GDD100 pKa = 3.16ILLYY104 pKa = 10.03GCNVARR110 pKa = 11.84DD111 pKa = 3.45QSGIDD116 pKa = 3.76FIGKK120 pKa = 8.99LAQLSGADD128 pKa = 3.14IAASNDD134 pKa = 3.26MTGSNTQNGDD144 pKa = 3.58WILEE148 pKa = 4.12SSIGMIDD155 pKa = 3.51TPALVLNQYY164 pKa = 9.56NQTLAAGTVTFTSSPSAATATDD186 pKa = 3.73GTASNDD192 pKa = 2.91IAGIVLNFTSDD203 pKa = 3.94LGTNWTYY210 pKa = 7.38EE211 pKa = 4.03TPYY214 pKa = 8.64TGSVITVGYY223 pKa = 7.85ATNAPQALMTIKK235 pKa = 10.7SSDD238 pKa = 3.19GSNFWFKK245 pKa = 11.42SLFIADD251 pKa = 3.83SGGQPVKK258 pKa = 11.24VEE260 pKa = 3.96GFEE263 pKa = 4.68NNVSTGSVNLDD274 pKa = 3.33TTGGGSLYY282 pKa = 10.67EE283 pKa = 4.19NTFGTASLTQSIFQNVDD300 pKa = 2.86EE301 pKa = 4.86VRR303 pKa = 11.84ITPQSGVQMWIGVNNIQIDD322 pKa = 4.12NPVLPGPTVTSALYY336 pKa = 10.22DD337 pKa = 3.4ASTNSLVVTGTNMTATAGVTNDD359 pKa = 2.9INVAKK364 pKa = 9.37LTLTGEE370 pKa = 4.37GGATYY375 pKa = 9.95TLTSSNVEE383 pKa = 3.73IDD385 pKa = 3.2SATQFTVALNATDD398 pKa = 3.69QINVEE403 pKa = 4.0GLLNKK408 pKa = 10.43NGTSSVGTTTYY419 pKa = 10.9NIAAAADD426 pKa = 3.5WDD428 pKa = 4.19VASTGNADD436 pKa = 3.44LTGNGITVSNTQTPTITSATYY457 pKa = 10.28DD458 pKa = 3.45AATGSLVVTGTNLVKK473 pKa = 8.89TTGATNDD480 pKa = 2.94ITVNKK485 pKa = 8.93LTFTGEE491 pKa = 4.0GGSTYY496 pKa = 10.6TLTSSNVEE504 pKa = 3.77ITDD507 pKa = 3.37ATSFSVTLNATDD519 pKa = 4.66LAALNQIVNKK529 pKa = 10.28NGTSSTGATTYY540 pKa = 11.38NLAAADD546 pKa = 4.09DD547 pKa = 4.34WNSVITAGTIADD559 pKa = 3.6ATSAVTASNVAVPAITNATYY579 pKa = 10.47DD580 pKa = 3.41ASTGALVVTGTGFLKK595 pKa = 10.96LSGATNDD602 pKa = 2.83IVANKK607 pKa = 8.26FTFTGEE613 pKa = 3.66GGATYY618 pKa = 10.2TLTDD622 pKa = 3.52TSNVEE627 pKa = 4.09ITSGTAFTITLSATDD642 pKa = 3.78KK643 pKa = 11.15AALNLMMDD651 pKa = 3.84KK652 pKa = 10.92NGTSSTSGTTYY663 pKa = 11.3NLAAAEE669 pKa = 4.06DD670 pKa = 3.91WAAGADD676 pKa = 3.67AAVVVADD683 pKa = 3.76TTGNGVTASNVAVPTITSSTYY704 pKa = 9.72NASTGALVVTGTGFLSKK721 pKa = 10.63SGATNDD727 pKa = 2.69IVANKK732 pKa = 8.26FTFTGEE738 pKa = 3.66GGATYY743 pKa = 10.2TLTDD747 pKa = 3.52TSNVEE752 pKa = 3.93ITSGTVFTITLSATDD767 pKa = 4.1LAGVDD772 pKa = 3.95QIMNKK777 pKa = 10.09NGTASTSGTTYY788 pKa = 11.3NLAAAEE794 pKa = 4.27DD795 pKa = 4.02WASGADD801 pKa = 3.49AAVVVADD808 pKa = 3.76TTGNGVTASNVAVPTITSATYY829 pKa = 10.15DD830 pKa = 3.54YY831 pKa = 8.69NTNVLTVTGTGFLNKK846 pKa = 10.07SGATNDD852 pKa = 2.97IDD854 pKa = 3.98LTKK857 pKa = 10.2LTFSGEE863 pKa = 4.13GGSTYY868 pKa = 10.77TLTNATGVEE877 pKa = 4.3ITSGTSFSVTLSGADD892 pKa = 3.96LINVEE897 pKa = 4.3TLLNKK902 pKa = 10.41NGTSAVSGTTYY913 pKa = 11.3NLAAAEE919 pKa = 4.06DD920 pKa = 3.91WAAGADD926 pKa = 3.67AAVVVADD933 pKa = 3.85TTGNGITLSNYY944 pKa = 8.44AAPTVTSATYY954 pKa = 10.34DD955 pKa = 3.04AGTHH959 pKa = 4.84VLSVTGTNLVSKK971 pKa = 10.72SGATNDD977 pKa = 3.49VAVSMLTVTGEE988 pKa = 3.96GGTYY992 pKa = 9.67TLTSSDD998 pKa = 3.36VDD1000 pKa = 3.25ITNATTFSVTLNAADD1015 pKa = 3.46QLVVNGLLNKK1025 pKa = 10.44NGTASSGATTYY1036 pKa = 11.46NLAAAEE1042 pKa = 4.14DD1043 pKa = 4.21WLAGTASATVVADD1056 pKa = 3.44LTGNGITVSNVQTPTITSATYY1077 pKa = 10.77DD1078 pKa = 3.64STTGSLVVTGTNFFSKK1094 pKa = 10.58VGANNDD1100 pKa = 2.86IDD1102 pKa = 4.22LTKK1105 pKa = 10.39LTFTGGAANATYY1117 pKa = 9.71TLSSTSNVEE1126 pKa = 3.67ITSATSFSITLSGADD1141 pKa = 3.58KK1142 pKa = 10.71TSVDD1146 pKa = 3.97ALLDD1150 pKa = 4.3NIGTTSSGGSTYY1162 pKa = 11.36NLAAANNWLAGADD1175 pKa = 3.61SAAVITDD1182 pKa = 3.74AVNAITVSINPAITSATYY1200 pKa = 10.2DD1201 pKa = 3.23ASTGSLVVTGTNMQVMAGAANDD1223 pKa = 3.19ITANKK1228 pKa = 8.7LTFTGEE1234 pKa = 3.65GGATYY1239 pKa = 10.15TLTDD1243 pKa = 3.5SANVEE1248 pKa = 4.17LSSATSFTIALSATDD1263 pKa = 3.83KK1264 pKa = 11.25AAINQIVNKK1273 pKa = 9.96NGTSSTGATTYY1284 pKa = 11.38NLAAADD1290 pKa = 3.73DD1291 pKa = 4.16WNAQVSAGDD1300 pKa = 3.57TSDD1303 pKa = 2.98ATNAVTVSSVAAPTITNATYY1323 pKa = 10.44DD1324 pKa = 3.59ASTGALVVTGTGFLKK1339 pKa = 10.96LSGATNDD1346 pKa = 2.83IVANKK1351 pKa = 8.26FTFTGEE1357 pKa = 3.66GGATYY1362 pKa = 10.2TLTDD1366 pKa = 3.52TSNVEE1371 pKa = 4.09ITSGTAFTITLSATDD1386 pKa = 3.78KK1387 pKa = 11.15AALNLMMDD1395 pKa = 3.84KK1396 pKa = 10.92NGTSSTSGTTYY1407 pKa = 11.3NLAAAEE1413 pKa = 4.06DD1414 pKa = 3.91WAAGADD1420 pKa = 3.67AAVVVADD1427 pKa = 3.76TTGNGVTASNVAVPTITSSTYY1448 pKa = 9.72NASTGALVVTGTGFLSKK1465 pKa = 10.63SGATNDD1471 pKa = 2.69IVANKK1476 pKa = 8.26FTFTGEE1482 pKa = 3.66GGATYY1487 pKa = 10.2TLTDD1491 pKa = 3.52TSNVEE1496 pKa = 3.93ITSGTVFTITLSATDD1511 pKa = 4.1LAGVDD1516 pKa = 3.95QIMNKK1521 pKa = 10.09NGTASTSGTTYY1532 pKa = 11.3NLAAAEE1538 pKa = 4.27DD1539 pKa = 4.02WASGADD1545 pKa = 3.49AAVVVADD1552 pKa = 3.76TTGNGVTASNVAVPAITSSTYY1573 pKa = 10.09NASTGALVVTGTGFLKK1589 pKa = 10.96LSGATNDD1596 pKa = 2.83IVANKK1601 pKa = 8.26FTFTGEE1607 pKa = 3.66GGATYY1612 pKa = 10.2TLTDD1616 pKa = 3.52TSNVEE1621 pKa = 4.09ITSGTAFTITLSATDD1636 pKa = 3.78KK1637 pKa = 11.15AALNLMMDD1645 pKa = 3.84KK1646 pKa = 10.92NGTSSTSGTTYY1657 pKa = 11.3NLAAAEE1663 pKa = 4.06DD1664 pKa = 3.91WAAGADD1670 pKa = 3.67AAVVVADD1677 pKa = 3.76TTGNGVTASNVAVPTITSSTYY1698 pKa = 9.72NASTGALVVTGTGFLSKK1715 pKa = 10.63SGATNDD1721 pKa = 2.69IVANKK1726 pKa = 8.26FTFTGEE1732 pKa = 3.66GGATYY1737 pKa = 10.2TLTDD1741 pKa = 3.52TSNVEE1746 pKa = 3.93ITSGTVFTITLSATDD1761 pKa = 4.1LAGVDD1766 pKa = 3.95QIMNKK1771 pKa = 10.09NGTASTSGTTYY1782 pKa = 11.3NLAAAEE1788 pKa = 4.27DD1789 pKa = 4.02WASGADD1795 pKa = 3.49AAVVVADD1802 pKa = 3.76TTGNGVTASNVAVPTITSATYY1823 pKa = 10.5DD1824 pKa = 3.25SGTGVLTVSGTHH1836 pKa = 5.88FVHH1839 pKa = 7.59KK1840 pKa = 10.46SGSTNDD1846 pKa = 3.44IDD1848 pKa = 6.19LSMLTFTGEE1857 pKa = 3.7AGGTYY1862 pKa = 9.82TLTTATDD1869 pKa = 3.53VEE1871 pKa = 4.73ITSGTSFTVTLSGADD1886 pKa = 3.31KK1887 pKa = 10.74TGVDD1891 pKa = 3.42TLLNKK1896 pKa = 10.11IGTSSTDD1903 pKa = 2.72ATTYY1907 pKa = 11.2NLAAAEE1913 pKa = 4.12DD1914 pKa = 4.16WATGADD1920 pKa = 3.26AAVNVVDD1927 pKa = 3.91ATNAITVSQTIASSGGGGGTSTPTTTIDD1955 pKa = 3.24GATVEE1960 pKa = 4.62TTTATNSSGQTVEE1973 pKa = 4.32QIIILPISSTRR1984 pKa = 11.84VNSTGDD1990 pKa = 3.05TATADD1995 pKa = 3.22IPLFWGEE2002 pKa = 3.87SSRR2005 pKa = 11.84TVDD2008 pKa = 2.86ATTANLPTGIGLSATGARR2026 pKa = 11.84APQSGHH2032 pKa = 6.45TIQDD2036 pKa = 3.83SINDD2040 pKa = 3.94LLALINEE2047 pKa = 4.78TAPSTDD2053 pKa = 3.04TSKK2056 pKa = 10.17TSMLGGGSTFLNTLSGITDD2075 pKa = 3.25NLVINKK2081 pKa = 7.94ITLTHH2086 pKa = 6.96TGTSAPGMPITIQGTTSTVTTSGGALPPVEE2116 pKa = 4.76ALVIDD2121 pKa = 4.47GQTLLSNTTLSLNNVEE2137 pKa = 4.28FATIIGEE2144 pKa = 4.0NLIIRR2149 pKa = 11.84GGEE2152 pKa = 3.79GRR2154 pKa = 11.84NTIFTGDD2161 pKa = 3.24GHH2163 pKa = 6.8QDD2165 pKa = 3.12IMCGTDD2171 pKa = 3.85DD2172 pKa = 5.74DD2173 pKa = 4.51EE2174 pKa = 4.61LHH2176 pKa = 7.08AGGGDD2181 pKa = 3.67DD2182 pKa = 3.82TVGSAGGNDD2191 pKa = 3.83RR2192 pKa = 11.84IFAEE2196 pKa = 4.64AGNDD2200 pKa = 3.54TVFGGTGDD2208 pKa = 4.27DD2209 pKa = 4.03FMHH2212 pKa = 6.75GGSGNDD2218 pKa = 2.96TVTYY2222 pKa = 10.2DD2223 pKa = 3.79GNRR2226 pKa = 11.84GDD2228 pKa = 4.28YY2229 pKa = 10.94VITRR2233 pKa = 11.84DD2234 pKa = 3.24NGKK2237 pKa = 7.74TYY2239 pKa = 10.75VYY2241 pKa = 9.47TVSTGEE2247 pKa = 3.8TDD2249 pKa = 3.49TVVNAEE2255 pKa = 4.49SIQFADD2261 pKa = 4.91AIYY2264 pKa = 9.56TITNNEE2270 pKa = 4.71DD2271 pKa = 3.02LTQIASLYY2279 pKa = 8.19QQILGRR2285 pKa = 11.84QADD2288 pKa = 4.02LDD2290 pKa = 3.88GFQYY2294 pKa = 7.64WTSTFDD2300 pKa = 3.27NGDD2303 pKa = 3.75TIGAIATSFVRR2314 pKa = 11.84SSEE2317 pKa = 4.06YY2318 pKa = 10.85FNDD2321 pKa = 3.7TGNVWDD2327 pKa = 5.19SMSNDD2332 pKa = 3.01QRR2334 pKa = 11.84IEE2336 pKa = 3.83TFYY2339 pKa = 11.42QLMLGRR2345 pKa = 11.84ASDD2348 pKa = 4.34AVGKK2352 pKa = 9.94AYY2354 pKa = 9.76WMDD2357 pKa = 5.06AINNGTMSIQDD2368 pKa = 3.25IAGSFVEE2375 pKa = 4.24SSEE2378 pKa = 3.83MQGIYY2383 pKa = 9.62TGQQAWSFFVV2393 pKa = 3.18
Molecular weight: 241.54 kDa
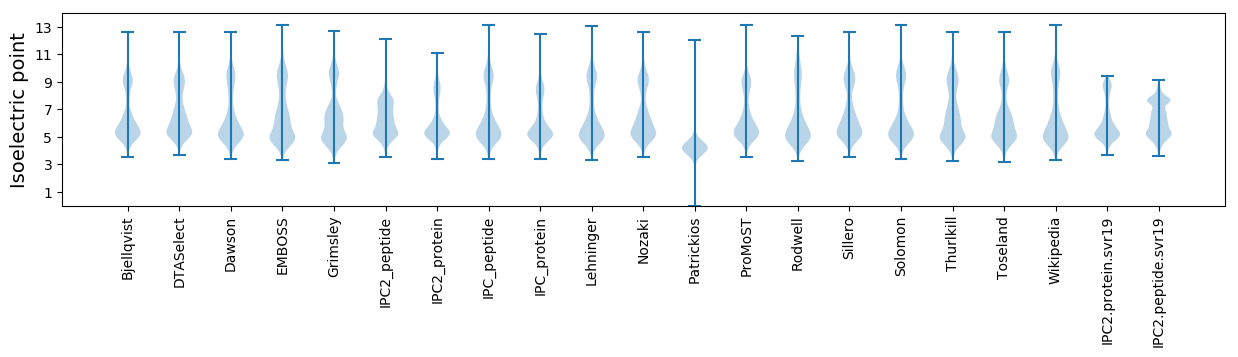
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4U0N6|E4U0N6_SULKY pEK499_p136 domain-containing protein OS=Sulfuricurvum kujiense (strain ATCC BAA-921 / DSM 16994 / JCM 11577 / YK-1) OX=709032 GN=Sulku_1692 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.0GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.15NGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.0GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.15NGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
867431 |
30 |
3343 |
310.5 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.729 ± 0.05 | 0.904 ± 0.017 |
5.551 ± 0.039 | 7.081 ± 0.047 |
4.584 ± 0.039 | 6.386 ± 0.05 |
2.313 ± 0.025 | 7.913 ± 0.043 |
6.467 ± 0.046 | 9.819 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.776 ± 0.026 | 4.539 ± 0.041 |
3.406 ± 0.03 | 3.225 ± 0.026 |
4.377 ± 0.034 | 6.79 ± 0.034 |
5.403 ± 0.056 | 6.186 ± 0.037 |
0.847 ± 0.015 | 3.705 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
