
Desulfocapsa sulfexigens (strain DSM 10523 / SB164P1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Desulfocapsa; Desulfocapsa sulfexigens
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
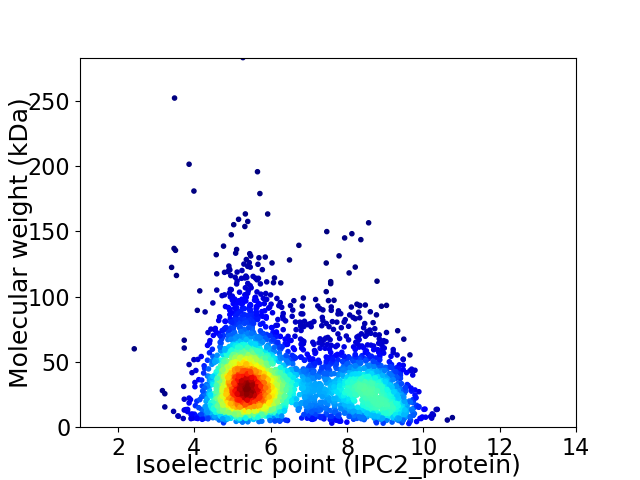
Virtual 2D-PAGE plot for 3514 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|M1PGR0|M1PGR0_DESSD 3-dehydroquinate synthase OS=Desulfocapsa sulfexigens (strain DSM 10523 / SB164P1) OX=1167006 GN=aroB PE=3 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84KK3 pKa = 9.21IVILVTCCFLLGFSNRR19 pKa = 11.84VHH21 pKa = 7.2ALEE24 pKa = 4.32ILYY27 pKa = 10.59NDD29 pKa = 4.37LSGGGINAIQMSAFNEE45 pKa = 3.99AASYY49 pKa = 9.31WEE51 pKa = 4.88SIFVDD56 pKa = 3.55NVTVRR61 pKa = 11.84VDD63 pKa = 3.3IDD65 pKa = 3.52MQALGTGIIGSAQSYY80 pKa = 9.93KK81 pKa = 10.29VDD83 pKa = 3.38TYY85 pKa = 11.71YY86 pKa = 11.68GDD88 pKa = 3.35IRR90 pKa = 11.84NALAGDD96 pKa = 3.68VRR98 pKa = 11.84STDD101 pKa = 4.78DD102 pKa = 4.43ILATSNLQTTDD113 pKa = 2.94HH114 pKa = 6.68LNVVVNDD121 pKa = 4.8LYY123 pKa = 11.39DD124 pKa = 5.18DD125 pKa = 3.52NHH127 pKa = 7.03AWILNSSSTVEE138 pKa = 4.19VNSEE142 pKa = 3.79LWVNRR147 pKa = 11.84ANLKK151 pKa = 10.31ALSLLNDD158 pKa = 4.73DD159 pKa = 4.58GQADD163 pKa = 3.95ASVLMSSNFSFDD175 pKa = 3.26YY176 pKa = 10.82DD177 pKa = 3.84RR178 pKa = 11.84SDD180 pKa = 5.52GIDD183 pKa = 3.41SDD185 pKa = 4.11KK186 pKa = 11.46LDD188 pKa = 3.63FAGVMVHH195 pKa = 7.03EE196 pKa = 5.38IGHH199 pKa = 6.34ALGFVSGVDD208 pKa = 2.84IMDD211 pKa = 3.62YY212 pKa = 10.98YY213 pKa = 11.58GLDD216 pKa = 3.65GPGNGSWDD224 pKa = 3.6WGWSGEE230 pKa = 4.17GSSNTLEE237 pKa = 4.25DD238 pKa = 2.85EE239 pKa = 5.21GYY241 pKa = 10.88FSVLDD246 pKa = 3.56LFRR249 pKa = 11.84YY250 pKa = 9.5SDD252 pKa = 4.14YY253 pKa = 11.12INEE256 pKa = 4.22YY257 pKa = 10.43EE258 pKa = 4.18SGSLFGYY265 pKa = 10.51LDD267 pKa = 3.18WTYY270 pKa = 11.45EE271 pKa = 4.2SVEE274 pKa = 4.29GGWGDD279 pKa = 3.73SYY281 pKa = 11.81FSFDD285 pKa = 4.37GGTTNLAPFSTGRR298 pKa = 11.84YY299 pKa = 9.31NGDD302 pKa = 3.52GYY304 pKa = 10.82QASHH308 pKa = 6.74WEE310 pKa = 4.13DD311 pKa = 3.59DD312 pKa = 3.86LGLGRR317 pKa = 11.84MDD319 pKa = 3.43PTTAYY324 pKa = 10.47GEE326 pKa = 4.45YY327 pKa = 10.49YY328 pKa = 10.83DD329 pKa = 5.78FLTPLDD335 pKa = 3.9LRR337 pKa = 11.84AFDD340 pKa = 4.13VIGWDD345 pKa = 3.97LSVTDD350 pKa = 5.1PVPEE354 pKa = 4.18PTTLLLFGAGLSGLIYY370 pKa = 10.33VRR372 pKa = 11.84SRR374 pKa = 11.84KK375 pKa = 9.1KK376 pKa = 10.48RR377 pKa = 11.84KK378 pKa = 9.18
MM1 pKa = 7.65RR2 pKa = 11.84KK3 pKa = 9.21IVILVTCCFLLGFSNRR19 pKa = 11.84VHH21 pKa = 7.2ALEE24 pKa = 4.32ILYY27 pKa = 10.59NDD29 pKa = 4.37LSGGGINAIQMSAFNEE45 pKa = 3.99AASYY49 pKa = 9.31WEE51 pKa = 4.88SIFVDD56 pKa = 3.55NVTVRR61 pKa = 11.84VDD63 pKa = 3.3IDD65 pKa = 3.52MQALGTGIIGSAQSYY80 pKa = 9.93KK81 pKa = 10.29VDD83 pKa = 3.38TYY85 pKa = 11.71YY86 pKa = 11.68GDD88 pKa = 3.35IRR90 pKa = 11.84NALAGDD96 pKa = 3.68VRR98 pKa = 11.84STDD101 pKa = 4.78DD102 pKa = 4.43ILATSNLQTTDD113 pKa = 2.94HH114 pKa = 6.68LNVVVNDD121 pKa = 4.8LYY123 pKa = 11.39DD124 pKa = 5.18DD125 pKa = 3.52NHH127 pKa = 7.03AWILNSSSTVEE138 pKa = 4.19VNSEE142 pKa = 3.79LWVNRR147 pKa = 11.84ANLKK151 pKa = 10.31ALSLLNDD158 pKa = 4.73DD159 pKa = 4.58GQADD163 pKa = 3.95ASVLMSSNFSFDD175 pKa = 3.26YY176 pKa = 10.82DD177 pKa = 3.84RR178 pKa = 11.84SDD180 pKa = 5.52GIDD183 pKa = 3.41SDD185 pKa = 4.11KK186 pKa = 11.46LDD188 pKa = 3.63FAGVMVHH195 pKa = 7.03EE196 pKa = 5.38IGHH199 pKa = 6.34ALGFVSGVDD208 pKa = 2.84IMDD211 pKa = 3.62YY212 pKa = 10.98YY213 pKa = 11.58GLDD216 pKa = 3.65GPGNGSWDD224 pKa = 3.6WGWSGEE230 pKa = 4.17GSSNTLEE237 pKa = 4.25DD238 pKa = 2.85EE239 pKa = 5.21GYY241 pKa = 10.88FSVLDD246 pKa = 3.56LFRR249 pKa = 11.84YY250 pKa = 9.5SDD252 pKa = 4.14YY253 pKa = 11.12INEE256 pKa = 4.22YY257 pKa = 10.43EE258 pKa = 4.18SGSLFGYY265 pKa = 10.51LDD267 pKa = 3.18WTYY270 pKa = 11.45EE271 pKa = 4.2SVEE274 pKa = 4.29GGWGDD279 pKa = 3.73SYY281 pKa = 11.81FSFDD285 pKa = 4.37GGTTNLAPFSTGRR298 pKa = 11.84YY299 pKa = 9.31NGDD302 pKa = 3.52GYY304 pKa = 10.82QASHH308 pKa = 6.74WEE310 pKa = 4.13DD311 pKa = 3.59DD312 pKa = 3.86LGLGRR317 pKa = 11.84MDD319 pKa = 3.43PTTAYY324 pKa = 10.47GEE326 pKa = 4.45YY327 pKa = 10.49YY328 pKa = 10.83DD329 pKa = 5.78FLTPLDD335 pKa = 3.9LRR337 pKa = 11.84AFDD340 pKa = 4.13VIGWDD345 pKa = 3.97LSVTDD350 pKa = 5.1PVPEE354 pKa = 4.18PTTLLLFGAGLSGLIYY370 pKa = 10.33VRR372 pKa = 11.84SRR374 pKa = 11.84KK375 pKa = 9.1KK376 pKa = 10.48RR377 pKa = 11.84KK378 pKa = 9.18
Molecular weight: 41.75 kDa
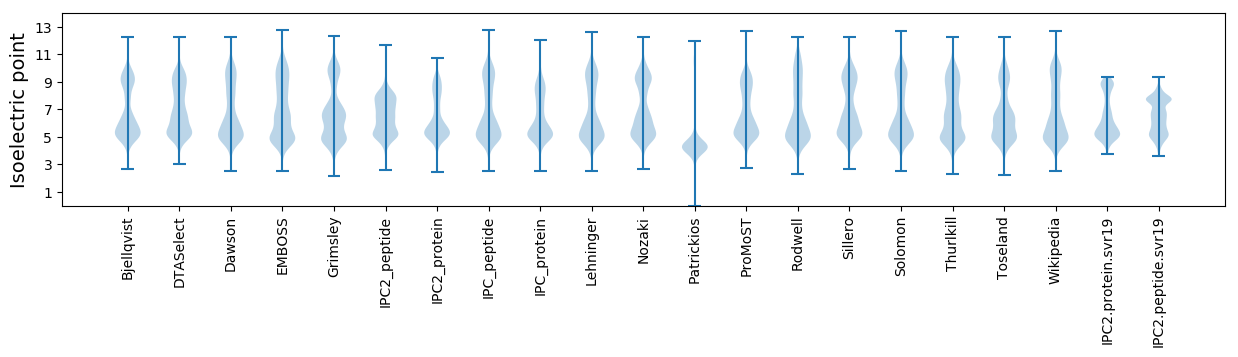
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1P2T4|M1P2T4_DESSD Uncharacterized protein OS=Desulfocapsa sulfexigens (strain DSM 10523 / SB164P1) OX=1167006 GN=UWK_01232 PE=4 SV=1
MM1 pKa = 7.59PPKK4 pKa = 10.37KK5 pKa = 9.98RR6 pKa = 11.84VFQRR10 pKa = 11.84KK11 pKa = 8.75RR12 pKa = 11.84SCRR15 pKa = 11.84FCSDD19 pKa = 2.86KK20 pKa = 11.1EE21 pKa = 3.78MSIDD25 pKa = 3.68YY26 pKa = 10.27KK27 pKa = 11.12DD28 pKa = 3.61PKK30 pKa = 8.85TLRR33 pKa = 11.84NFVTEE38 pKa = 4.04RR39 pKa = 11.84GKK41 pKa = 10.51IIPRR45 pKa = 11.84RR46 pKa = 11.84IYY48 pKa = 8.18GTCATHH54 pKa = 6.63QRR56 pKa = 11.84QLCEE60 pKa = 3.94AIKK63 pKa = 10.25RR64 pKa = 11.84ARR66 pKa = 11.84QIALLPYY73 pKa = 10.19SGSTQRR79 pKa = 4.61
MM1 pKa = 7.59PPKK4 pKa = 10.37KK5 pKa = 9.98RR6 pKa = 11.84VFQRR10 pKa = 11.84KK11 pKa = 8.75RR12 pKa = 11.84SCRR15 pKa = 11.84FCSDD19 pKa = 2.86KK20 pKa = 11.1EE21 pKa = 3.78MSIDD25 pKa = 3.68YY26 pKa = 10.27KK27 pKa = 11.12DD28 pKa = 3.61PKK30 pKa = 8.85TLRR33 pKa = 11.84NFVTEE38 pKa = 4.04RR39 pKa = 11.84GKK41 pKa = 10.51IIPRR45 pKa = 11.84RR46 pKa = 11.84IYY48 pKa = 8.18GTCATHH54 pKa = 6.63QRR56 pKa = 11.84QLCEE60 pKa = 3.94AIKK63 pKa = 10.25RR64 pKa = 11.84ARR66 pKa = 11.84QIALLPYY73 pKa = 10.19SGSTQRR79 pKa = 4.61
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190136 |
23 |
2577 |
338.7 |
37.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.674 ± 0.037 | 1.329 ± 0.015 |
5.493 ± 0.041 | 6.508 ± 0.036 |
4.471 ± 0.027 | 7.155 ± 0.04 |
2.172 ± 0.019 | 7.018 ± 0.029 |
5.719 ± 0.04 | 10.483 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.595 ± 0.018 | 3.946 ± 0.025 |
4.075 ± 0.025 | 3.685 ± 0.028 |
5.006 ± 0.033 | 6.571 ± 0.036 |
5.461 ± 0.026 | 6.689 ± 0.033 |
1.018 ± 0.013 | 2.932 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
