
Mytilus galloprovincialis (Mediterranean mussel)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Bivalvia; Autobranchia; Pteriomorphia; Mytiloida; Mytiloidea; Mytilidae; Mytilinae; Mytilus
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
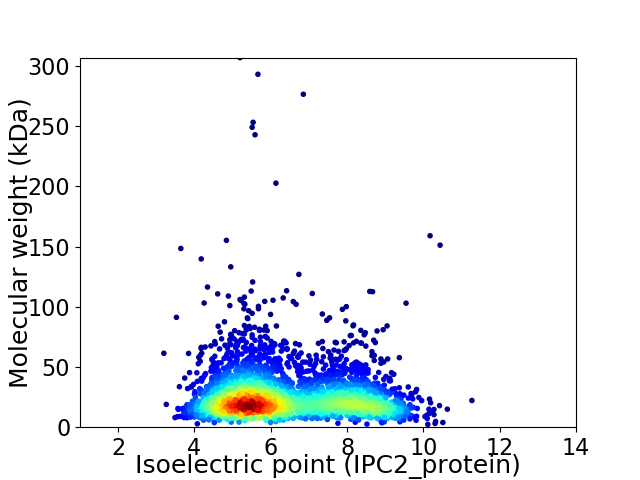
Virtual 2D-PAGE plot for 3147 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L5TR72|A0A3L5TR72_MYTGA Sodium-transporter and chloride-dependent neurotransmitter (Fragment) OS=Mytilus galloprovincialis OX=29158 GN=AM593_02716 PE=4 SV=1
MM1 pKa = 7.22KK2 pKa = 10.33RR3 pKa = 11.84KK4 pKa = 9.28ILHH7 pKa = 6.51RR8 pKa = 11.84KK9 pKa = 9.23LKK11 pKa = 9.77TEE13 pKa = 3.87QHH15 pKa = 5.1EE16 pKa = 4.46HH17 pKa = 4.62NQKK20 pKa = 10.66LGLISMTFEE29 pKa = 4.33GQCKK33 pKa = 9.21FGRR36 pKa = 11.84RR37 pKa = 11.84NYY39 pKa = 9.8VLKK42 pKa = 10.48QASQLGVARR51 pKa = 11.84QLDD54 pKa = 4.06TMVTLCRR61 pKa = 11.84FQPDD65 pKa = 3.12VTIQIDD71 pKa = 4.11RR72 pKa = 11.84LPALAMVYY80 pKa = 10.21CRR82 pKa = 11.84TEE84 pKa = 3.91DD85 pKa = 3.74VITVAQSSSTIDD97 pKa = 4.36PITEE101 pKa = 4.7IITVPPIIPIILDD114 pKa = 3.59DD115 pKa = 4.1VLLSGALVRR124 pKa = 11.84DD125 pKa = 3.76LLYY128 pKa = 11.16SNKK131 pKa = 9.64KK132 pKa = 9.49DD133 pKa = 3.19INVIISKK140 pKa = 10.09EE141 pKa = 3.92ILSASLDD148 pKa = 3.59VDD150 pKa = 4.17DD151 pKa = 4.34VCPRR155 pKa = 11.84SVDD158 pKa = 3.66DD159 pKa = 3.82VCVKK163 pKa = 10.5FVDD166 pKa = 4.22AVCAGFVDD174 pKa = 4.87DD175 pKa = 4.04VCVGYY180 pKa = 10.38FEE182 pKa = 5.06VVCTRR187 pKa = 11.84FVDD190 pKa = 4.61DD191 pKa = 3.43VCKK194 pKa = 10.57RR195 pKa = 11.84LLNDD199 pKa = 2.88VCTVFVDD206 pKa = 4.39DD207 pKa = 3.9VCAGFVDD214 pKa = 5.89DD215 pKa = 3.81VCAWFVDD222 pKa = 4.02DD223 pKa = 5.05VFAGFIANVCAEE235 pKa = 4.4FVDD238 pKa = 4.25DD239 pKa = 4.62LCAGFVDD246 pKa = 4.87DD247 pKa = 4.19VCAGFVDD254 pKa = 4.73DD255 pKa = 4.14VCAGFVDD262 pKa = 4.73DD263 pKa = 3.97VCAGFVVDD271 pKa = 3.82VCAGFVDD278 pKa = 4.73DD279 pKa = 4.14VCAGFVDD286 pKa = 4.51KK287 pKa = 11.1VWAGFVDD294 pKa = 5.98DD295 pKa = 3.89VCTWFDD301 pKa = 3.66DD302 pKa = 4.39FVFAGFVDD310 pKa = 4.65DD311 pKa = 3.97VCAGFVDD318 pKa = 4.73DD319 pKa = 4.14VCAGFVDD326 pKa = 4.73DD327 pKa = 4.14VCAGFVDD334 pKa = 4.73DD335 pKa = 3.97VCAGFVVDD343 pKa = 3.82VCAGFVDD350 pKa = 4.73DD351 pKa = 4.14VCAGFVDD358 pKa = 4.51KK359 pKa = 11.1VWAGFVDD366 pKa = 4.87DD367 pKa = 5.35GLLMMYY373 pKa = 9.93VQGLLMMSAQGLLMISAHH391 pKa = 6.53ALLMMSAHH399 pKa = 7.05ALLMMSAHH407 pKa = 6.41GLLMISAQGLL417 pKa = 3.6
MM1 pKa = 7.22KK2 pKa = 10.33RR3 pKa = 11.84KK4 pKa = 9.28ILHH7 pKa = 6.51RR8 pKa = 11.84KK9 pKa = 9.23LKK11 pKa = 9.77TEE13 pKa = 3.87QHH15 pKa = 5.1EE16 pKa = 4.46HH17 pKa = 4.62NQKK20 pKa = 10.66LGLISMTFEE29 pKa = 4.33GQCKK33 pKa = 9.21FGRR36 pKa = 11.84RR37 pKa = 11.84NYY39 pKa = 9.8VLKK42 pKa = 10.48QASQLGVARR51 pKa = 11.84QLDD54 pKa = 4.06TMVTLCRR61 pKa = 11.84FQPDD65 pKa = 3.12VTIQIDD71 pKa = 4.11RR72 pKa = 11.84LPALAMVYY80 pKa = 10.21CRR82 pKa = 11.84TEE84 pKa = 3.91DD85 pKa = 3.74VITVAQSSSTIDD97 pKa = 4.36PITEE101 pKa = 4.7IITVPPIIPIILDD114 pKa = 3.59DD115 pKa = 4.1VLLSGALVRR124 pKa = 11.84DD125 pKa = 3.76LLYY128 pKa = 11.16SNKK131 pKa = 9.64KK132 pKa = 9.49DD133 pKa = 3.19INVIISKK140 pKa = 10.09EE141 pKa = 3.92ILSASLDD148 pKa = 3.59VDD150 pKa = 4.17DD151 pKa = 4.34VCPRR155 pKa = 11.84SVDD158 pKa = 3.66DD159 pKa = 3.82VCVKK163 pKa = 10.5FVDD166 pKa = 4.22AVCAGFVDD174 pKa = 4.87DD175 pKa = 4.04VCVGYY180 pKa = 10.38FEE182 pKa = 5.06VVCTRR187 pKa = 11.84FVDD190 pKa = 4.61DD191 pKa = 3.43VCKK194 pKa = 10.57RR195 pKa = 11.84LLNDD199 pKa = 2.88VCTVFVDD206 pKa = 4.39DD207 pKa = 3.9VCAGFVDD214 pKa = 5.89DD215 pKa = 3.81VCAWFVDD222 pKa = 4.02DD223 pKa = 5.05VFAGFIANVCAEE235 pKa = 4.4FVDD238 pKa = 4.25DD239 pKa = 4.62LCAGFVDD246 pKa = 4.87DD247 pKa = 4.19VCAGFVDD254 pKa = 4.73DD255 pKa = 4.14VCAGFVDD262 pKa = 4.73DD263 pKa = 3.97VCAGFVVDD271 pKa = 3.82VCAGFVDD278 pKa = 4.73DD279 pKa = 4.14VCAGFVDD286 pKa = 4.51KK287 pKa = 11.1VWAGFVDD294 pKa = 5.98DD295 pKa = 3.89VCTWFDD301 pKa = 3.66DD302 pKa = 4.39FVFAGFVDD310 pKa = 4.65DD311 pKa = 3.97VCAGFVDD318 pKa = 4.73DD319 pKa = 4.14VCAGFVDD326 pKa = 4.73DD327 pKa = 4.14VCAGFVDD334 pKa = 4.73DD335 pKa = 3.97VCAGFVVDD343 pKa = 3.82VCAGFVDD350 pKa = 4.73DD351 pKa = 4.14VCAGFVDD358 pKa = 4.51KK359 pKa = 11.1VWAGFVDD366 pKa = 4.87DD367 pKa = 5.35GLLMMYY373 pKa = 9.93VQGLLMMSAQGLLMISAHH391 pKa = 6.53ALLMMSAHH399 pKa = 7.05ALLMMSAHH407 pKa = 6.41GLLMISAQGLL417 pKa = 3.6
Molecular weight: 45.3 kDa
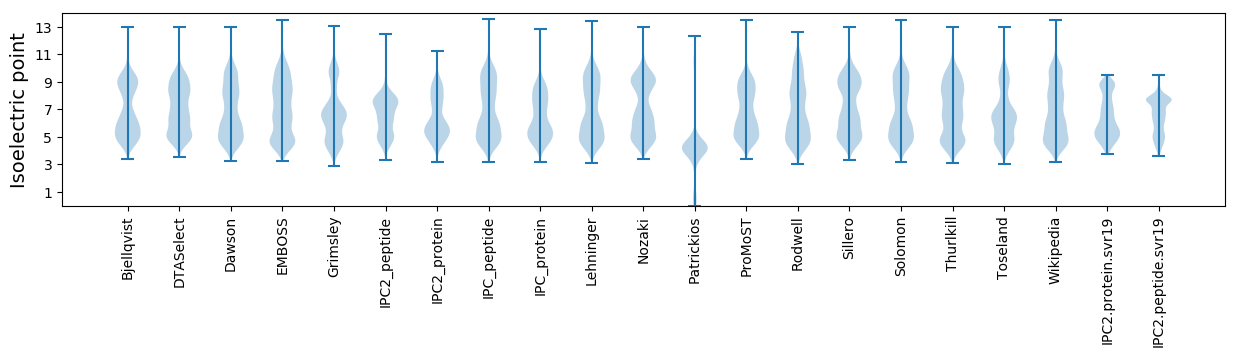
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L5TT46|A0A3L5TT46_MYTGA Uncharacterized protein (Fragment) OS=Mytilus galloprovincialis OX=29158 GN=AM593_08727 PE=4 SV=1
MM1 pKa = 7.6AVPAKK6 pKa = 10.04IVLKK10 pKa = 9.72STTKK14 pKa = 10.0MSLNDD19 pKa = 3.68RR20 pKa = 11.84FSTIQATVRR29 pKa = 11.84QPAQQASVSNIRR41 pKa = 11.84AKK43 pKa = 9.82MAAQHH48 pKa = 5.74QATAANKK55 pKa = 9.21RR56 pKa = 11.84LAMQMANRR64 pKa = 11.84PSVQAALKK72 pKa = 10.09IKK74 pKa = 10.36KK75 pKa = 9.7KK76 pKa = 9.28SLKK79 pKa = 9.71QRR81 pKa = 11.84LGFGNTSVKK90 pKa = 10.5SRR92 pKa = 11.84LTLGGRR98 pKa = 11.84GGGAIVRR105 pKa = 11.84GGRR108 pKa = 11.84GQGRR112 pKa = 11.84LMRR115 pKa = 11.84GRR117 pKa = 11.84GQMRR121 pKa = 11.84GRR123 pKa = 11.84GRR125 pKa = 11.84GGRR128 pKa = 11.84GGSGSTPGSPTGNQMVRR145 pKa = 11.84GGNRR149 pKa = 11.84GQRR152 pKa = 11.84GGFVQRR158 pKa = 11.84GRR160 pKa = 11.84GQRR163 pKa = 11.84GGFVQRR169 pKa = 11.84GEE171 pKa = 3.81NRR173 pKa = 11.84GRR175 pKa = 11.84RR176 pKa = 11.84GGMQRR181 pKa = 11.84GRR183 pKa = 11.84GFQRR187 pKa = 11.84GNRR190 pKa = 11.84GGRR193 pKa = 11.84GQTQGFRR200 pKa = 11.84GNRR203 pKa = 11.84GSGGFRR209 pKa = 3.24
MM1 pKa = 7.6AVPAKK6 pKa = 10.04IVLKK10 pKa = 9.72STTKK14 pKa = 10.0MSLNDD19 pKa = 3.68RR20 pKa = 11.84FSTIQATVRR29 pKa = 11.84QPAQQASVSNIRR41 pKa = 11.84AKK43 pKa = 9.82MAAQHH48 pKa = 5.74QATAANKK55 pKa = 9.21RR56 pKa = 11.84LAMQMANRR64 pKa = 11.84PSVQAALKK72 pKa = 10.09IKK74 pKa = 10.36KK75 pKa = 9.7KK76 pKa = 9.28SLKK79 pKa = 9.71QRR81 pKa = 11.84LGFGNTSVKK90 pKa = 10.5SRR92 pKa = 11.84LTLGGRR98 pKa = 11.84GGGAIVRR105 pKa = 11.84GGRR108 pKa = 11.84GQGRR112 pKa = 11.84LMRR115 pKa = 11.84GRR117 pKa = 11.84GQMRR121 pKa = 11.84GRR123 pKa = 11.84GRR125 pKa = 11.84GGRR128 pKa = 11.84GGSGSTPGSPTGNQMVRR145 pKa = 11.84GGNRR149 pKa = 11.84GQRR152 pKa = 11.84GGFVQRR158 pKa = 11.84GRR160 pKa = 11.84GQRR163 pKa = 11.84GGFVQRR169 pKa = 11.84GEE171 pKa = 3.81NRR173 pKa = 11.84GRR175 pKa = 11.84RR176 pKa = 11.84GGMQRR181 pKa = 11.84GRR183 pKa = 11.84GFQRR187 pKa = 11.84GNRR190 pKa = 11.84GGRR193 pKa = 11.84GQTQGFRR200 pKa = 11.84GNRR203 pKa = 11.84GSGGFRR209 pKa = 3.24
Molecular weight: 22.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
761033 |
18 |
2702 |
241.8 |
27.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.877 ± 0.036 | 2.443 ± 0.043 |
6.143 ± 0.047 | 6.574 ± 0.054 |
3.878 ± 0.034 | 5.624 ± 0.069 |
2.417 ± 0.025 | 6.442 ± 0.047 |
7.278 ± 0.061 | 8.234 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.021 | 5.551 ± 0.042 |
4.145 ± 0.048 | 4.186 ± 0.041 |
4.515 ± 0.038 | 7.787 ± 0.048 |
6.738 ± 0.092 | 6.227 ± 0.039 |
1.079 ± 0.017 | 3.445 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
