
Apis mellifera associated microvirus 14
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
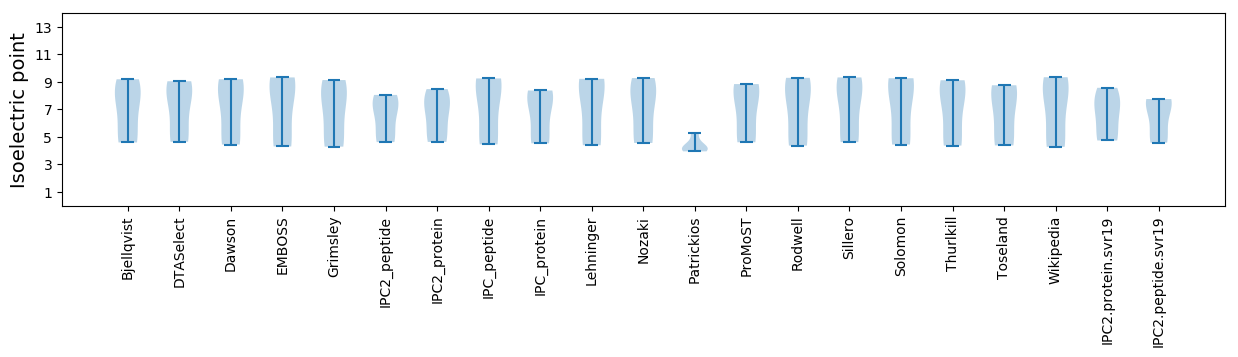
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTY1|A0A3S8UTY1_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 14 OX=2494741 PE=4 SV=1
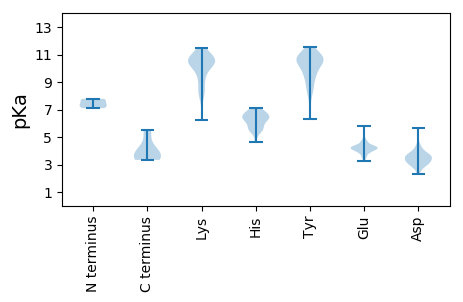
MM1 pKa = 7.78DD2 pKa = 5.62AEE4 pKa = 4.2TGEE7 pKa = 4.3IIEE10 pKa = 4.7RR11 pKa = 11.84KK12 pKa = 9.41PPFIRR17 pKa = 11.84TAYY20 pKa = 8.45NYY22 pKa = 10.88SMEE25 pKa = 4.37AVSEE29 pKa = 4.3EE30 pKa = 4.23TGLTCPEE37 pKa = 4.23DD38 pKa = 3.58SRR40 pKa = 11.84TKK42 pKa = 10.63QEE44 pKa = 4.63FKK46 pKa = 11.24DD47 pKa = 3.49EE48 pKa = 3.98ADD50 pKa = 2.98INKK53 pKa = 9.29IVEE56 pKa = 4.25RR57 pKa = 11.84FGLTGEE63 pKa = 4.63LPQNLRR69 pKa = 11.84MPISGDD75 pKa = 3.29FTGLGDD81 pKa = 3.48YY82 pKa = 10.56HH83 pKa = 6.75EE84 pKa = 4.99AMNLVLQAQEE94 pKa = 4.72EE95 pKa = 4.46FDD97 pKa = 3.76KK98 pKa = 11.48LPAKK102 pKa = 10.16VRR104 pKa = 11.84EE105 pKa = 4.19RR106 pKa = 11.84FGNDD110 pKa = 2.32AGAFIAWAEE119 pKa = 4.09RR120 pKa = 11.84EE121 pKa = 4.31DD122 pKa = 3.86NLEE125 pKa = 3.88EE126 pKa = 4.38AKK128 pKa = 10.98LLGIARR134 pKa = 11.84EE135 pKa = 4.36EE136 pKa = 4.22KK137 pKa = 10.67AKK139 pKa = 9.81PQPIEE144 pKa = 3.56VRR146 pKa = 11.84LAKK149 pKa = 10.66EE150 pKa = 3.96EE151 pKa = 4.25TPPPAA156 pKa = 5.48
MM1 pKa = 7.78DD2 pKa = 5.62AEE4 pKa = 4.2TGEE7 pKa = 4.3IIEE10 pKa = 4.7RR11 pKa = 11.84KK12 pKa = 9.41PPFIRR17 pKa = 11.84TAYY20 pKa = 8.45NYY22 pKa = 10.88SMEE25 pKa = 4.37AVSEE29 pKa = 4.3EE30 pKa = 4.23TGLTCPEE37 pKa = 4.23DD38 pKa = 3.58SRR40 pKa = 11.84TKK42 pKa = 10.63QEE44 pKa = 4.63FKK46 pKa = 11.24DD47 pKa = 3.49EE48 pKa = 3.98ADD50 pKa = 2.98INKK53 pKa = 9.29IVEE56 pKa = 4.25RR57 pKa = 11.84FGLTGEE63 pKa = 4.63LPQNLRR69 pKa = 11.84MPISGDD75 pKa = 3.29FTGLGDD81 pKa = 3.48YY82 pKa = 10.56HH83 pKa = 6.75EE84 pKa = 4.99AMNLVLQAQEE94 pKa = 4.72EE95 pKa = 4.46FDD97 pKa = 3.76KK98 pKa = 11.48LPAKK102 pKa = 10.16VRR104 pKa = 11.84EE105 pKa = 4.19RR106 pKa = 11.84FGNDD110 pKa = 2.32AGAFIAWAEE119 pKa = 4.09RR120 pKa = 11.84EE121 pKa = 4.31DD122 pKa = 3.86NLEE125 pKa = 3.88EE126 pKa = 4.38AKK128 pKa = 10.98LLGIARR134 pKa = 11.84EE135 pKa = 4.36EE136 pKa = 4.22KK137 pKa = 10.67AKK139 pKa = 9.81PQPIEE144 pKa = 3.56VRR146 pKa = 11.84LAKK149 pKa = 10.66EE150 pKa = 3.96EE151 pKa = 4.25TPPPAA156 pKa = 5.48
Molecular weight: 17.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U605|A0A3Q8U605_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 14 OX=2494741 PE=4 SV=1
MM1 pKa = 7.71PCYY4 pKa = 10.38NPLKK8 pKa = 10.39AYY10 pKa = 10.33QLTNGEE16 pKa = 4.32IKK18 pKa = 10.37FHH20 pKa = 5.36EE21 pKa = 4.31TRR23 pKa = 11.84NVQRR27 pKa = 11.84EE28 pKa = 4.15LKK30 pKa = 9.99LPCGQCIGCRR40 pKa = 11.84LEE42 pKa = 5.77RR43 pKa = 11.84SRR45 pKa = 11.84QWAVRR50 pKa = 11.84CMHH53 pKa = 6.81EE54 pKa = 4.19ARR56 pKa = 11.84MYY58 pKa = 9.21EE59 pKa = 4.11QNCFITLTYY68 pKa = 10.62APEE71 pKa = 4.19HH72 pKa = 5.47VPKK75 pKa = 10.51RR76 pKa = 11.84GSLDD80 pKa = 3.15YY81 pKa = 11.42SVFQKK86 pKa = 10.52FMKK89 pKa = 10.35RR90 pKa = 11.84LRR92 pKa = 11.84KK93 pKa = 8.18HH94 pKa = 5.65TGPEE98 pKa = 3.23RR99 pKa = 11.84VRR101 pKa = 11.84FYY103 pKa = 10.48MCGEE107 pKa = 4.04YY108 pKa = 10.94GEE110 pKa = 4.82TEE112 pKa = 4.25HH113 pKa = 7.05RR114 pKa = 11.84PHH116 pKa = 6.28YY117 pKa = 9.47HH118 pKa = 6.2ACLFGYY124 pKa = 9.6DD125 pKa = 3.69WPDD128 pKa = 2.64KK129 pKa = 10.44TFFRR133 pKa = 11.84TTPTGEE139 pKa = 3.61KK140 pKa = 9.94LYY142 pKa = 10.73RR143 pKa = 11.84SEE145 pKa = 4.2TLEE148 pKa = 4.02KK149 pKa = 9.99LWPYY153 pKa = 11.16GFATTGTLTFEE164 pKa = 4.03SAAYY168 pKa = 8.37VARR171 pKa = 11.84YY172 pKa = 6.32CTKK175 pKa = 10.71KK176 pKa = 9.16ITGHH180 pKa = 5.53NAKK183 pKa = 10.15DD184 pKa = 3.05HH185 pKa = 6.23YY186 pKa = 10.54KK187 pKa = 10.3RR188 pKa = 11.84QDD190 pKa = 3.07AEE192 pKa = 4.3GEE194 pKa = 4.36YY195 pKa = 10.48EE196 pKa = 4.1LEE198 pKa = 4.26PEE200 pKa = 4.5FGHH203 pKa = 6.56MSLKK207 pKa = 10.43PGIGAAWLAKK217 pKa = 10.3YY218 pKa = 10.61GNTDD222 pKa = 2.77VWPFDD227 pKa = 3.7HH228 pKa = 7.12VITRR232 pKa = 11.84GMEE235 pKa = 4.17TKK237 pKa = 10.12PPKK240 pKa = 10.79YY241 pKa = 9.5YY242 pKa = 10.91DD243 pKa = 4.43KK244 pKa = 10.69ILKK247 pKa = 8.13RR248 pKa = 11.84TNPDD252 pKa = 2.52RR253 pKa = 11.84FAEE256 pKa = 4.17IQEE259 pKa = 4.26SRR261 pKa = 11.84EE262 pKa = 3.62VTAAQWAADD271 pKa = 3.7QTPDD275 pKa = 3.05RR276 pKa = 11.84LAAKK280 pKa = 8.83EE281 pKa = 3.95QVKK284 pKa = 9.93RR285 pKa = 11.84AQLRR289 pKa = 11.84QLKK292 pKa = 8.43RR293 pKa = 11.84TLL295 pKa = 3.34
MM1 pKa = 7.71PCYY4 pKa = 10.38NPLKK8 pKa = 10.39AYY10 pKa = 10.33QLTNGEE16 pKa = 4.32IKK18 pKa = 10.37FHH20 pKa = 5.36EE21 pKa = 4.31TRR23 pKa = 11.84NVQRR27 pKa = 11.84EE28 pKa = 4.15LKK30 pKa = 9.99LPCGQCIGCRR40 pKa = 11.84LEE42 pKa = 5.77RR43 pKa = 11.84SRR45 pKa = 11.84QWAVRR50 pKa = 11.84CMHH53 pKa = 6.81EE54 pKa = 4.19ARR56 pKa = 11.84MYY58 pKa = 9.21EE59 pKa = 4.11QNCFITLTYY68 pKa = 10.62APEE71 pKa = 4.19HH72 pKa = 5.47VPKK75 pKa = 10.51RR76 pKa = 11.84GSLDD80 pKa = 3.15YY81 pKa = 11.42SVFQKK86 pKa = 10.52FMKK89 pKa = 10.35RR90 pKa = 11.84LRR92 pKa = 11.84KK93 pKa = 8.18HH94 pKa = 5.65TGPEE98 pKa = 3.23RR99 pKa = 11.84VRR101 pKa = 11.84FYY103 pKa = 10.48MCGEE107 pKa = 4.04YY108 pKa = 10.94GEE110 pKa = 4.82TEE112 pKa = 4.25HH113 pKa = 7.05RR114 pKa = 11.84PHH116 pKa = 6.28YY117 pKa = 9.47HH118 pKa = 6.2ACLFGYY124 pKa = 9.6DD125 pKa = 3.69WPDD128 pKa = 2.64KK129 pKa = 10.44TFFRR133 pKa = 11.84TTPTGEE139 pKa = 3.61KK140 pKa = 9.94LYY142 pKa = 10.73RR143 pKa = 11.84SEE145 pKa = 4.2TLEE148 pKa = 4.02KK149 pKa = 9.99LWPYY153 pKa = 11.16GFATTGTLTFEE164 pKa = 4.03SAAYY168 pKa = 8.37VARR171 pKa = 11.84YY172 pKa = 6.32CTKK175 pKa = 10.71KK176 pKa = 9.16ITGHH180 pKa = 5.53NAKK183 pKa = 10.15DD184 pKa = 3.05HH185 pKa = 6.23YY186 pKa = 10.54KK187 pKa = 10.3RR188 pKa = 11.84QDD190 pKa = 3.07AEE192 pKa = 4.3GEE194 pKa = 4.36YY195 pKa = 10.48EE196 pKa = 4.1LEE198 pKa = 4.26PEE200 pKa = 4.5FGHH203 pKa = 6.56MSLKK207 pKa = 10.43PGIGAAWLAKK217 pKa = 10.3YY218 pKa = 10.61GNTDD222 pKa = 2.77VWPFDD227 pKa = 3.7HH228 pKa = 7.12VITRR232 pKa = 11.84GMEE235 pKa = 4.17TKK237 pKa = 10.12PPKK240 pKa = 10.79YY241 pKa = 9.5YY242 pKa = 10.91DD243 pKa = 4.43KK244 pKa = 10.69ILKK247 pKa = 8.13RR248 pKa = 11.84TNPDD252 pKa = 2.52RR253 pKa = 11.84FAEE256 pKa = 4.17IQEE259 pKa = 4.26SRR261 pKa = 11.84EE262 pKa = 3.62VTAAQWAADD271 pKa = 3.7QTPDD275 pKa = 3.05RR276 pKa = 11.84LAAKK280 pKa = 8.83EE281 pKa = 3.95QVKK284 pKa = 9.93RR285 pKa = 11.84AQLRR289 pKa = 11.84QLKK292 pKa = 8.43RR293 pKa = 11.84TLL295 pKa = 3.34
Molecular weight: 34.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1364 |
81 |
534 |
272.8 |
30.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.63 ± 1.22 | 0.953 ± 0.581 |
4.692 ± 0.301 | 7.185 ± 2.177 |
4.106 ± 0.602 | 6.891 ± 0.703 |
2.126 ± 0.713 | 4.839 ± 0.584 |
4.545 ± 1.052 | 6.672 ± 0.23 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.372 ± 0.343 | 4.839 ± 0.935 |
5.352 ± 1.109 | 4.912 ± 0.738 |
7.185 ± 1.031 | 4.472 ± 0.869 |
7.698 ± 0.991 | 4.179 ± 0.67 |
1.393 ± 0.226 | 3.959 ± 1.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
