
Vicingus serpentipes
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Vicingaceae; Vicingus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
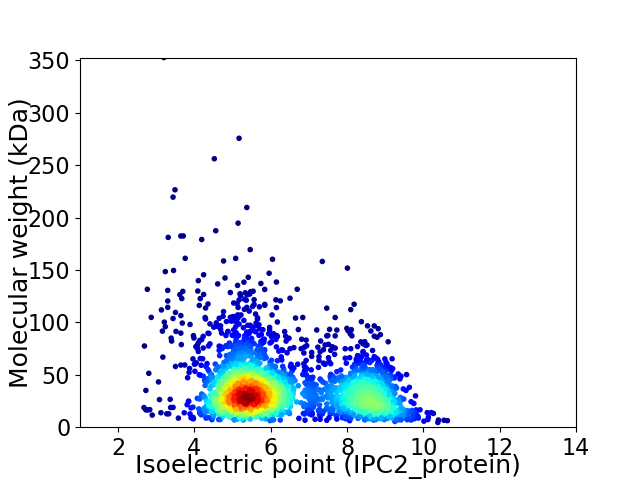
Virtual 2D-PAGE plot for 2518 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
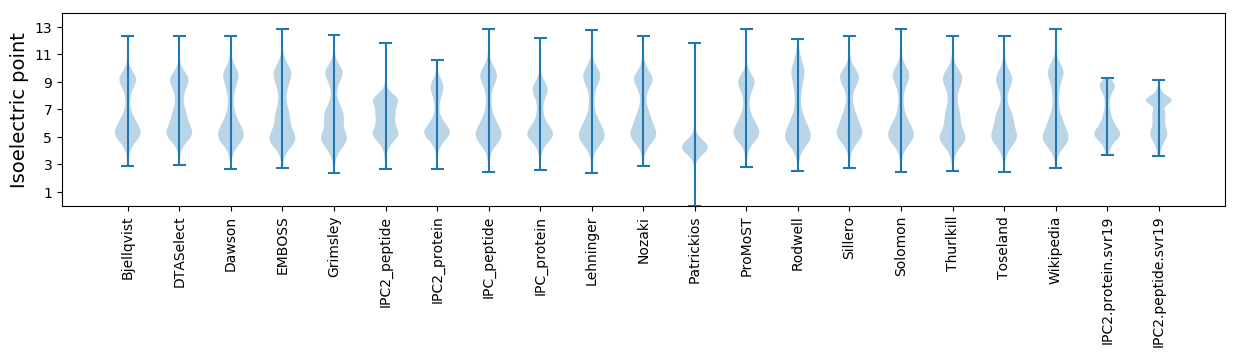
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6RWW8|A0A5C6RWW8_9FLAO CoA transferase OS=Vicingus serpentipes OX=1926625 GN=FRY74_00430 PE=4 SV=1
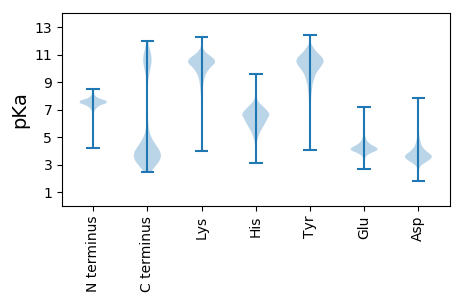
MM1 pKa = 7.71KK2 pKa = 10.27SYY4 pKa = 10.89LKK6 pKa = 10.57QIFLFATLIVTNSALAQQLDD26 pKa = 3.75WLGHH30 pKa = 4.95FRR32 pKa = 11.84STNHH36 pKa = 5.44VKK38 pKa = 9.04STAISTDD45 pKa = 2.85NDD47 pKa = 3.47GNIYY51 pKa = 10.35VVGTIEE57 pKa = 5.37IDD59 pKa = 3.84SADD62 pKa = 4.28LNPDD66 pKa = 2.76AGTYY70 pKa = 9.39YY71 pKa = 10.96LSPEE75 pKa = 4.11FSNAAFVCKK84 pKa = 10.1LDD86 pKa = 3.79SLGNFIWAKK95 pKa = 10.06SFGGGSSISPTSVAADD111 pKa = 3.64DD112 pKa = 4.1FGGVYY117 pKa = 10.04IGGSYY122 pKa = 10.6FNTVDD127 pKa = 4.42FDD129 pKa = 4.45PGLGVYY135 pKa = 10.43DD136 pKa = 3.38LTVFGGSDD144 pKa = 3.49LFLSKK149 pKa = 11.13LNVNGDD155 pKa = 3.76FEE157 pKa = 4.64WAFNAGSTSTDD168 pKa = 2.93AVTDD172 pKa = 3.53IAVDD176 pKa = 3.26IDD178 pKa = 3.81NNVYY182 pKa = 8.05ITGYY186 pKa = 10.42FDD188 pKa = 4.45YY189 pKa = 11.54ADD191 pKa = 4.55FDD193 pKa = 4.52PGAGWFALNPPEE205 pKa = 4.04KK206 pKa = 10.08AGYY209 pKa = 7.58IAKK212 pKa = 9.79YY213 pKa = 9.92KK214 pKa = 10.17PDD216 pKa = 3.41GSFVNAVDD224 pKa = 4.34FEE226 pKa = 5.49GDD228 pKa = 3.58GPSGSNSIAIAPNGSLYY245 pKa = 11.0VVGAFNSTIDD255 pKa = 3.62FDD257 pKa = 4.58PGVGTFNLTSISSNVDD273 pKa = 2.71IYY275 pKa = 11.38VCKK278 pKa = 10.48LDD280 pKa = 4.04TSLTFQWAKK289 pKa = 10.96SFGSSSADD297 pKa = 3.17NGHH300 pKa = 5.84TVSVSDD306 pKa = 3.61SDD308 pKa = 3.92EE309 pKa = 4.0PVITGGFWNTIDD321 pKa = 4.04VDD323 pKa = 4.37PGPSTYY329 pKa = 11.1NVTSNGLVDD338 pKa = 4.62NLLLKK343 pKa = 10.66LDD345 pKa = 3.8ASGNFMWANTYY356 pKa = 11.06GNAQTEE362 pKa = 4.39YY363 pKa = 11.03SLDD366 pKa = 3.26AAIAVNDD373 pKa = 4.91DD374 pKa = 3.34IYY376 pKa = 11.09ILGKK380 pKa = 10.43FIDD383 pKa = 4.84SIDD386 pKa = 3.56VDD388 pKa = 4.31GGPGTQLIASVSPSDD403 pKa = 4.03SYY405 pKa = 11.09IARR408 pKa = 11.84LDD410 pKa = 3.45GSGNLISSGQLGGPGSVSGKK430 pKa = 9.28GLCISNTQKK439 pKa = 10.32IYY441 pKa = 9.14HH442 pKa = 6.17TGIFSGSAEE451 pKa = 4.62LDD453 pKa = 3.3PSAGIDD459 pKa = 3.51SLTSLGLNDD468 pKa = 4.68SYY470 pKa = 11.4IQIAIPSAMVGLKK483 pKa = 9.54TISTEE488 pKa = 4.12TTKK491 pKa = 10.95KK492 pKa = 10.35EE493 pKa = 3.97LIKK496 pKa = 10.65IIDD499 pKa = 3.14IMGRR503 pKa = 11.84YY504 pKa = 9.26IKK506 pKa = 10.07DD507 pKa = 3.39QPNTLLIYY515 pKa = 10.19VYY517 pKa = 11.21SDD519 pKa = 2.97GTTEE523 pKa = 3.76KK524 pKa = 10.55VFRR527 pKa = 11.84VEE529 pKa = 3.78
MM1 pKa = 7.71KK2 pKa = 10.27SYY4 pKa = 10.89LKK6 pKa = 10.57QIFLFATLIVTNSALAQQLDD26 pKa = 3.75WLGHH30 pKa = 4.95FRR32 pKa = 11.84STNHH36 pKa = 5.44VKK38 pKa = 9.04STAISTDD45 pKa = 2.85NDD47 pKa = 3.47GNIYY51 pKa = 10.35VVGTIEE57 pKa = 5.37IDD59 pKa = 3.84SADD62 pKa = 4.28LNPDD66 pKa = 2.76AGTYY70 pKa = 9.39YY71 pKa = 10.96LSPEE75 pKa = 4.11FSNAAFVCKK84 pKa = 10.1LDD86 pKa = 3.79SLGNFIWAKK95 pKa = 10.06SFGGGSSISPTSVAADD111 pKa = 3.64DD112 pKa = 4.1FGGVYY117 pKa = 10.04IGGSYY122 pKa = 10.6FNTVDD127 pKa = 4.42FDD129 pKa = 4.45PGLGVYY135 pKa = 10.43DD136 pKa = 3.38LTVFGGSDD144 pKa = 3.49LFLSKK149 pKa = 11.13LNVNGDD155 pKa = 3.76FEE157 pKa = 4.64WAFNAGSTSTDD168 pKa = 2.93AVTDD172 pKa = 3.53IAVDD176 pKa = 3.26IDD178 pKa = 3.81NNVYY182 pKa = 8.05ITGYY186 pKa = 10.42FDD188 pKa = 4.45YY189 pKa = 11.54ADD191 pKa = 4.55FDD193 pKa = 4.52PGAGWFALNPPEE205 pKa = 4.04KK206 pKa = 10.08AGYY209 pKa = 7.58IAKK212 pKa = 9.79YY213 pKa = 9.92KK214 pKa = 10.17PDD216 pKa = 3.41GSFVNAVDD224 pKa = 4.34FEE226 pKa = 5.49GDD228 pKa = 3.58GPSGSNSIAIAPNGSLYY245 pKa = 11.0VVGAFNSTIDD255 pKa = 3.62FDD257 pKa = 4.58PGVGTFNLTSISSNVDD273 pKa = 2.71IYY275 pKa = 11.38VCKK278 pKa = 10.48LDD280 pKa = 4.04TSLTFQWAKK289 pKa = 10.96SFGSSSADD297 pKa = 3.17NGHH300 pKa = 5.84TVSVSDD306 pKa = 3.61SDD308 pKa = 3.92EE309 pKa = 4.0PVITGGFWNTIDD321 pKa = 4.04VDD323 pKa = 4.37PGPSTYY329 pKa = 11.1NVTSNGLVDD338 pKa = 4.62NLLLKK343 pKa = 10.66LDD345 pKa = 3.8ASGNFMWANTYY356 pKa = 11.06GNAQTEE362 pKa = 4.39YY363 pKa = 11.03SLDD366 pKa = 3.26AAIAVNDD373 pKa = 4.91DD374 pKa = 3.34IYY376 pKa = 11.09ILGKK380 pKa = 10.43FIDD383 pKa = 4.84SIDD386 pKa = 3.56VDD388 pKa = 4.31GGPGTQLIASVSPSDD403 pKa = 4.03SYY405 pKa = 11.09IARR408 pKa = 11.84LDD410 pKa = 3.45GSGNLISSGQLGGPGSVSGKK430 pKa = 9.28GLCISNTQKK439 pKa = 10.32IYY441 pKa = 9.14HH442 pKa = 6.17TGIFSGSAEE451 pKa = 4.62LDD453 pKa = 3.3PSAGIDD459 pKa = 3.51SLTSLGLNDD468 pKa = 4.68SYY470 pKa = 11.4IQIAIPSAMVGLKK483 pKa = 9.54TISTEE488 pKa = 4.12TTKK491 pKa = 10.95KK492 pKa = 10.35EE493 pKa = 3.97LIKK496 pKa = 10.65IIDD499 pKa = 3.14IMGRR503 pKa = 11.84YY504 pKa = 9.26IKK506 pKa = 10.07DD507 pKa = 3.39QPNTLLIYY515 pKa = 10.19VYY517 pKa = 11.21SDD519 pKa = 2.97GTTEE523 pKa = 3.76KK524 pKa = 10.55VFRR527 pKa = 11.84VEE529 pKa = 3.78
Molecular weight: 56.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6S045|A0A5C6S045_9FLAO DUF368 domain-containing protein OS=Vicingus serpentipes OX=1926625 GN=FRY74_02015 PE=4 SV=1
MM1 pKa = 7.5AKK3 pKa = 10.25EE4 pKa = 3.94SMKK7 pKa = 10.47ARR9 pKa = 11.84EE10 pKa = 4.29RR11 pKa = 11.84KK12 pKa = 9.4RR13 pKa = 11.84EE14 pKa = 4.0AMVEE18 pKa = 3.95KK19 pKa = 10.81YY20 pKa = 10.44ADD22 pKa = 3.02KK23 pKa = 10.81RR24 pKa = 11.84AALKK28 pKa = 10.63AAGDD32 pKa = 3.42WDD34 pKa = 4.21ALQKK38 pKa = 10.78LPKK41 pKa = 9.8NSSKK45 pKa = 10.38VRR47 pKa = 11.84LHH49 pKa = 5.96NRR51 pKa = 11.84CKK53 pKa = 9.95LTGRR57 pKa = 11.84PRR59 pKa = 11.84GYY61 pKa = 9.01MRR63 pKa = 11.84QFGISRR69 pKa = 11.84VTFRR73 pKa = 11.84EE74 pKa = 3.92MASAGLIPGIKK85 pKa = 8.99KK86 pKa = 10.68ASWW89 pKa = 2.94
MM1 pKa = 7.5AKK3 pKa = 10.25EE4 pKa = 3.94SMKK7 pKa = 10.47ARR9 pKa = 11.84EE10 pKa = 4.29RR11 pKa = 11.84KK12 pKa = 9.4RR13 pKa = 11.84EE14 pKa = 4.0AMVEE18 pKa = 3.95KK19 pKa = 10.81YY20 pKa = 10.44ADD22 pKa = 3.02KK23 pKa = 10.81RR24 pKa = 11.84AALKK28 pKa = 10.63AAGDD32 pKa = 3.42WDD34 pKa = 4.21ALQKK38 pKa = 10.78LPKK41 pKa = 9.8NSSKK45 pKa = 10.38VRR47 pKa = 11.84LHH49 pKa = 5.96NRR51 pKa = 11.84CKK53 pKa = 9.95LTGRR57 pKa = 11.84PRR59 pKa = 11.84GYY61 pKa = 9.01MRR63 pKa = 11.84QFGISRR69 pKa = 11.84VTFRR73 pKa = 11.84EE74 pKa = 3.92MASAGLIPGIKK85 pKa = 8.99KK86 pKa = 10.68ASWW89 pKa = 2.94
Molecular weight: 10.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
911875 |
38 |
3426 |
362.1 |
40.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.045 ± 0.055 | 0.91 ± 0.023 |
5.535 ± 0.04 | 6.593 ± 0.065 |
5.168 ± 0.045 | 6.5 ± 0.06 |
1.6 ± 0.022 | 8.521 ± 0.05 |
7.922 ± 0.086 | 9.039 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.046 ± 0.025 | 6.885 ± 0.047 |
3.277 ± 0.032 | 3.152 ± 0.026 |
2.815 ± 0.036 | 6.774 ± 0.046 |
5.844 ± 0.077 | 6.155 ± 0.038 |
0.987 ± 0.016 | 4.231 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
