
Barley yellow dwarf virus (isolate PAV) (BYDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins
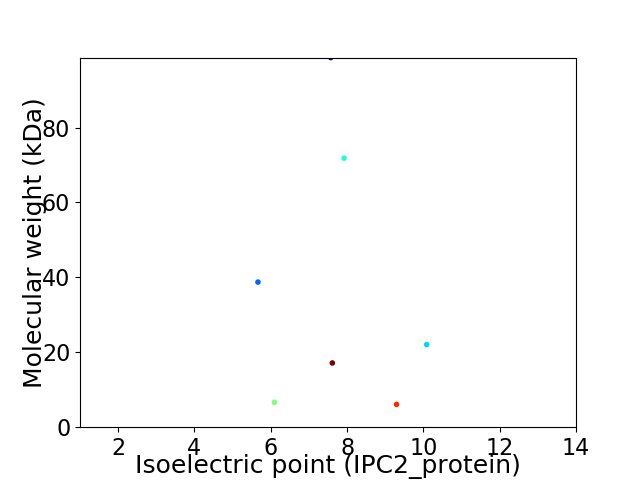
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P09505-2|RDRP-2_BYDVP Isoform of P09505 Isoform 39 kDa protein of Putative RNA-directed RNA polymerase OS=Barley yellow dwarf virus (isolate PAV) OX=2169986 GN=ORF1/ORF2 PE=4 SV=2
MM1 pKa = 7.29FFEE4 pKa = 4.37ILIGASAKK12 pKa = 9.9AVKK15 pKa = 10.39DD16 pKa = 4.76FISHH20 pKa = 6.66CYY22 pKa = 10.06SRR24 pKa = 11.84LKK26 pKa = 10.37SIYY29 pKa = 9.71YY30 pKa = 9.79SFKK33 pKa = 9.13RR34 pKa = 11.84WLMEE38 pKa = 3.49ISGQFKK44 pKa = 10.55AHH46 pKa = 6.75DD47 pKa = 4.2AFVNMCFGHH56 pKa = 6.08MADD59 pKa = 4.13IEE61 pKa = 4.26DD62 pKa = 4.48FEE64 pKa = 6.67AEE66 pKa = 3.94LAEE69 pKa = 4.22EE70 pKa = 4.04FAEE73 pKa = 4.79RR74 pKa = 11.84EE75 pKa = 4.3DD76 pKa = 3.93EE77 pKa = 4.31VEE79 pKa = 4.12EE80 pKa = 4.29ARR82 pKa = 11.84SLLKK86 pKa = 10.96LLVAQKK92 pKa = 10.25SKK94 pKa = 11.19SGVTEE99 pKa = 3.65AWTDD103 pKa = 3.69FFTKK107 pKa = 10.38SRR109 pKa = 11.84GGVYY113 pKa = 10.38APLSCEE119 pKa = 3.84PTRR122 pKa = 11.84QEE124 pKa = 5.16LEE126 pKa = 4.27VKK128 pKa = 10.02SEE130 pKa = 3.81KK131 pKa = 10.67LEE133 pKa = 4.14RR134 pKa = 11.84LLEE137 pKa = 4.24EE138 pKa = 3.56QHH140 pKa = 5.56QFEE143 pKa = 4.32VRR145 pKa = 11.84AAKK148 pKa = 10.1KK149 pKa = 9.89YY150 pKa = 9.82IKK152 pKa = 10.21EE153 pKa = 3.96KK154 pKa = 10.45GRR156 pKa = 11.84GFINCWNDD164 pKa = 2.75LRR166 pKa = 11.84SRR168 pKa = 11.84LRR170 pKa = 11.84LVKK173 pKa = 10.26DD174 pKa = 3.8VKK176 pKa = 11.28DD177 pKa = 3.65EE178 pKa = 4.5AKK180 pKa = 10.96DD181 pKa = 3.6NARR184 pKa = 11.84AAAKK188 pKa = 10.08IGAEE192 pKa = 3.9MFAPVDD198 pKa = 3.85VQDD201 pKa = 4.83LYY203 pKa = 12.01SFTEE207 pKa = 4.12VKK209 pKa = 10.42KK210 pKa = 11.31VEE212 pKa = 4.03TGLMKK217 pKa = 10.55EE218 pKa = 4.31VVKK221 pKa = 10.66EE222 pKa = 4.01KK223 pKa = 10.85NGEE226 pKa = 4.13EE227 pKa = 4.15EE228 pKa = 4.05KK229 pKa = 10.79HH230 pKa = 6.15LEE232 pKa = 4.42PIMEE236 pKa = 4.39EE237 pKa = 3.79VRR239 pKa = 11.84SIKK242 pKa = 9.8DD243 pKa = 3.03TAEE246 pKa = 3.95ARR248 pKa = 11.84DD249 pKa = 3.8AASTWITEE257 pKa = 4.33TVKK260 pKa = 11.0LKK262 pKa = 10.83NATLNADD269 pKa = 4.14EE270 pKa = 5.48LSLATIARR278 pKa = 11.84YY279 pKa = 9.55VEE281 pKa = 3.98NVGDD285 pKa = 4.18KK286 pKa = 10.66FKK288 pKa = 11.42LDD290 pKa = 3.69IASKK294 pKa = 9.49TYY296 pKa = 10.25LKK298 pKa = 10.28QVASMSVPIPTNKK311 pKa = 10.06DD312 pKa = 2.74IKK314 pKa = 10.87LKK316 pKa = 10.16MVLQSPEE323 pKa = 3.1ARR325 pKa = 11.84ARR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.79RR330 pKa = 11.84MDD332 pKa = 3.34VLDD335 pKa = 3.87SVGFF339 pKa = 3.75
MM1 pKa = 7.29FFEE4 pKa = 4.37ILIGASAKK12 pKa = 9.9AVKK15 pKa = 10.39DD16 pKa = 4.76FISHH20 pKa = 6.66CYY22 pKa = 10.06SRR24 pKa = 11.84LKK26 pKa = 10.37SIYY29 pKa = 9.71YY30 pKa = 9.79SFKK33 pKa = 9.13RR34 pKa = 11.84WLMEE38 pKa = 3.49ISGQFKK44 pKa = 10.55AHH46 pKa = 6.75DD47 pKa = 4.2AFVNMCFGHH56 pKa = 6.08MADD59 pKa = 4.13IEE61 pKa = 4.26DD62 pKa = 4.48FEE64 pKa = 6.67AEE66 pKa = 3.94LAEE69 pKa = 4.22EE70 pKa = 4.04FAEE73 pKa = 4.79RR74 pKa = 11.84EE75 pKa = 4.3DD76 pKa = 3.93EE77 pKa = 4.31VEE79 pKa = 4.12EE80 pKa = 4.29ARR82 pKa = 11.84SLLKK86 pKa = 10.96LLVAQKK92 pKa = 10.25SKK94 pKa = 11.19SGVTEE99 pKa = 3.65AWTDD103 pKa = 3.69FFTKK107 pKa = 10.38SRR109 pKa = 11.84GGVYY113 pKa = 10.38APLSCEE119 pKa = 3.84PTRR122 pKa = 11.84QEE124 pKa = 5.16LEE126 pKa = 4.27VKK128 pKa = 10.02SEE130 pKa = 3.81KK131 pKa = 10.67LEE133 pKa = 4.14RR134 pKa = 11.84LLEE137 pKa = 4.24EE138 pKa = 3.56QHH140 pKa = 5.56QFEE143 pKa = 4.32VRR145 pKa = 11.84AAKK148 pKa = 10.1KK149 pKa = 9.89YY150 pKa = 9.82IKK152 pKa = 10.21EE153 pKa = 3.96KK154 pKa = 10.45GRR156 pKa = 11.84GFINCWNDD164 pKa = 2.75LRR166 pKa = 11.84SRR168 pKa = 11.84LRR170 pKa = 11.84LVKK173 pKa = 10.26DD174 pKa = 3.8VKK176 pKa = 11.28DD177 pKa = 3.65EE178 pKa = 4.5AKK180 pKa = 10.96DD181 pKa = 3.6NARR184 pKa = 11.84AAAKK188 pKa = 10.08IGAEE192 pKa = 3.9MFAPVDD198 pKa = 3.85VQDD201 pKa = 4.83LYY203 pKa = 12.01SFTEE207 pKa = 4.12VKK209 pKa = 10.42KK210 pKa = 11.31VEE212 pKa = 4.03TGLMKK217 pKa = 10.55EE218 pKa = 4.31VVKK221 pKa = 10.66EE222 pKa = 4.01KK223 pKa = 10.85NGEE226 pKa = 4.13EE227 pKa = 4.15EE228 pKa = 4.05KK229 pKa = 10.79HH230 pKa = 6.15LEE232 pKa = 4.42PIMEE236 pKa = 4.39EE237 pKa = 3.79VRR239 pKa = 11.84SIKK242 pKa = 9.8DD243 pKa = 3.03TAEE246 pKa = 3.95ARR248 pKa = 11.84DD249 pKa = 3.8AASTWITEE257 pKa = 4.33TVKK260 pKa = 11.0LKK262 pKa = 10.83NATLNADD269 pKa = 4.14EE270 pKa = 5.48LSLATIARR278 pKa = 11.84YY279 pKa = 9.55VEE281 pKa = 3.98NVGDD285 pKa = 4.18KK286 pKa = 10.66FKK288 pKa = 11.42LDD290 pKa = 3.69IASKK294 pKa = 9.49TYY296 pKa = 10.25LKK298 pKa = 10.28QVASMSVPIPTNKK311 pKa = 10.06DD312 pKa = 2.74IKK314 pKa = 10.87LKK316 pKa = 10.16MVLQSPEE323 pKa = 3.1ARR325 pKa = 11.84ARR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.79RR330 pKa = 11.84MDD332 pKa = 3.34VLDD335 pKa = 3.87SVGFF339 pKa = 3.75
Molecular weight: 38.74 kDa
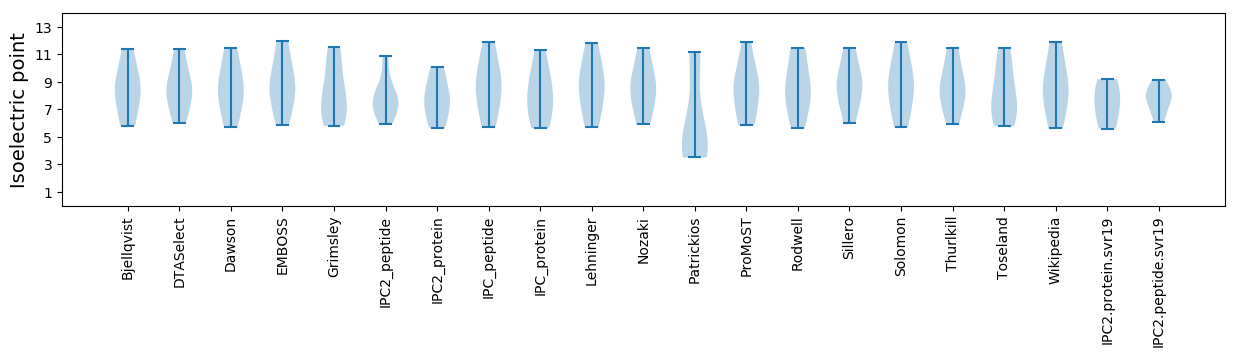
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P09513|MVP_BYDVP Movement protein OS=Barley yellow dwarf virus (isolate PAV) OX=2169986 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.32NSVGRR6 pKa = 11.84RR7 pKa = 11.84GPRR10 pKa = 11.84RR11 pKa = 11.84ANQNGTRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TVRR26 pKa = 11.84PVVVVQPNRR35 pKa = 11.84AGPRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84NGRR44 pKa = 11.84RR45 pKa = 11.84KK46 pKa = 10.36GRR48 pKa = 11.84GGANFVFRR56 pKa = 11.84PTGGTEE62 pKa = 3.86VFVFSVDD69 pKa = 3.29NLKK72 pKa = 11.06ANSSGAIKK80 pKa = 10.42FGPSLSQCPALSDD93 pKa = 4.84GILKK97 pKa = 10.33SYY99 pKa = 10.27HH100 pKa = 6.5RR101 pKa = 11.84YY102 pKa = 9.53KK103 pKa = 9.44ITSIRR108 pKa = 11.84VEE110 pKa = 4.62FKK112 pKa = 10.49SHH114 pKa = 6.49ASANTAGAIFIEE126 pKa = 5.16LDD128 pKa = 3.7TACKK132 pKa = 9.92QSALGSYY139 pKa = 9.83INSFTISKK147 pKa = 8.05TASKK151 pKa = 9.21TFRR154 pKa = 11.84SEE156 pKa = 4.17AINGKK161 pKa = 7.99EE162 pKa = 4.03FQEE165 pKa = 4.27STIDD169 pKa = 3.81QFWMLYY175 pKa = 8.9KK176 pKa = 10.97ANGTTTDD183 pKa = 3.17TAGQFIITMSVSLMTAKK200 pKa = 10.59
MM1 pKa = 7.32NSVGRR6 pKa = 11.84RR7 pKa = 11.84GPRR10 pKa = 11.84RR11 pKa = 11.84ANQNGTRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TVRR26 pKa = 11.84PVVVVQPNRR35 pKa = 11.84AGPRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84NGRR44 pKa = 11.84RR45 pKa = 11.84KK46 pKa = 10.36GRR48 pKa = 11.84GGANFVFRR56 pKa = 11.84PTGGTEE62 pKa = 3.86VFVFSVDD69 pKa = 3.29NLKK72 pKa = 11.06ANSSGAIKK80 pKa = 10.42FGPSLSQCPALSDD93 pKa = 4.84GILKK97 pKa = 10.33SYY99 pKa = 10.27HH100 pKa = 6.5RR101 pKa = 11.84YY102 pKa = 9.53KK103 pKa = 9.44ITSIRR108 pKa = 11.84VEE110 pKa = 4.62FKK112 pKa = 10.49SHH114 pKa = 6.49ASANTAGAIFIEE126 pKa = 5.16LDD128 pKa = 3.7TACKK132 pKa = 9.92QSALGSYY139 pKa = 9.83INSFTISKK147 pKa = 8.05TASKK151 pKa = 9.21TFRR154 pKa = 11.84SEE156 pKa = 4.17AINGKK161 pKa = 7.99EE162 pKa = 4.03FQEE165 pKa = 4.27STIDD169 pKa = 3.81QFWMLYY175 pKa = 8.9KK176 pKa = 10.97ANGTTTDD183 pKa = 3.17TAGQFIITMSVSLMTAKK200 pKa = 10.59
Molecular weight: 22.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2327 |
55 |
867 |
332.4 |
37.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.778 ± 0.49 | 1.418 ± 0.251 |
4.641 ± 0.348 | 7.434 ± 0.866 |
4.083 ± 0.381 | 5.887 ± 0.468 |
1.676 ± 0.316 | 5.329 ± 0.274 |
7.134 ± 0.998 | 7.005 ± 0.865 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.578 ± 0.3 | 4.211 ± 0.402 |
4.856 ± 1.04 | 3.997 ± 0.677 |
6.489 ± 0.591 | 8.337 ± 0.675 |
6.231 ± 1.011 | 6.661 ± 0.393 |
0.945 ± 0.095 | 3.309 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
