
Methanococcus voltae (strain ATCC BAA-1334 / A3)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanococci; Methanococcales; Methanococcaceae; Methanococcus; Methanococcus voltae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
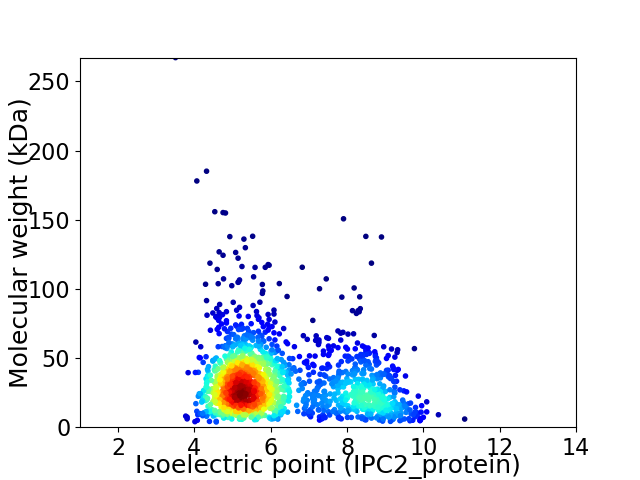
Virtual 2D-PAGE plot for 1658 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7DR98|D7DR98_METV3 Flagellin OS=Methanococcus voltae (strain ATCC BAA-1334 / A3) OX=456320 GN=Mvol_1720 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.41NNNILKK8 pKa = 9.99IFLCVMVSLVVMCSGCVSSSSSDD31 pKa = 3.41SADD34 pKa = 3.05DD35 pKa = 3.64VVVEE39 pKa = 4.45KK40 pKa = 10.42TSTNDD45 pKa = 2.94NTEE48 pKa = 3.81PKK50 pKa = 10.21EE51 pKa = 4.06NEE53 pKa = 3.79EE54 pKa = 4.35TKK56 pKa = 10.86DD57 pKa = 3.48DD58 pKa = 4.16EE59 pKa = 4.51KK60 pKa = 11.22PKK62 pKa = 10.07TEE64 pKa = 4.02EE65 pKa = 3.9PVEE68 pKa = 4.13EE69 pKa = 4.53GLPDD73 pKa = 3.32EE74 pKa = 4.55YY75 pKa = 11.56VAVFYY80 pKa = 10.15EE81 pKa = 4.59AYY83 pKa = 8.33STKK86 pKa = 10.64SFDD89 pKa = 3.16GSYY92 pKa = 10.82GKK94 pKa = 10.65EE95 pKa = 3.43EE96 pKa = 3.88AADD99 pKa = 3.68GKK101 pKa = 10.92KK102 pKa = 10.63YY103 pKa = 10.71LIVDD107 pKa = 3.56ITVYY111 pKa = 11.17NNGYY115 pKa = 10.52DD116 pKa = 3.9EE117 pKa = 5.33ISVNPLYY124 pKa = 10.75FDD126 pKa = 5.56AIVDD130 pKa = 4.01GVEE133 pKa = 3.57YY134 pKa = 10.2DD135 pKa = 3.8YY136 pKa = 11.6SIDD139 pKa = 4.09SYY141 pKa = 11.77SLEE144 pKa = 4.03NDD146 pKa = 3.16LDD148 pKa = 4.26SVNLRR153 pKa = 11.84NGGKK157 pKa = 8.49TSGSLAFEE165 pKa = 4.28IPEE168 pKa = 4.84DD169 pKa = 3.78SSSTYY174 pKa = 10.44SIEE177 pKa = 4.1YY178 pKa = 10.58NPITWDD184 pKa = 3.43KK185 pKa = 11.53LNIIYY190 pKa = 10.59NPEE193 pKa = 3.93DD194 pKa = 3.51VDD196 pKa = 4.04IEE198 pKa = 4.25EE199 pKa = 4.37AQKK202 pKa = 10.99AAASSSNTGAEE213 pKa = 3.93KK214 pKa = 9.82STPTTSNDD222 pKa = 3.34DD223 pKa = 3.62TDD225 pKa = 4.07SDD227 pKa = 4.3SVNIITAATVTEE239 pKa = 4.19SFNYY243 pKa = 10.53NIGSYY248 pKa = 9.91EE249 pKa = 3.96YY250 pKa = 10.27TRR252 pKa = 11.84EE253 pKa = 4.08APSGKK258 pKa = 8.63TYY260 pKa = 10.76LITYY264 pKa = 10.17ISIEE268 pKa = 3.94NNGYY272 pKa = 10.5DD273 pKa = 4.35EE274 pKa = 5.39ISTSPLYY281 pKa = 10.63FDD283 pKa = 6.1AIVDD287 pKa = 4.11GVGYY291 pKa = 10.33SYY293 pKa = 11.73ASDD296 pKa = 3.52TYY298 pKa = 11.27SLEE301 pKa = 3.93NHH303 pKa = 6.55LEE305 pKa = 4.15SVDD308 pKa = 3.6LRR310 pKa = 11.84NGGKK314 pKa = 8.47TSGLLIYY321 pKa = 7.7EE322 pKa = 4.23IPKK325 pKa = 10.81GSDD328 pKa = 3.18SYY330 pKa = 11.26EE331 pKa = 4.09LEE333 pKa = 4.15YY334 pKa = 11.3DD335 pKa = 4.17PISWDD340 pKa = 3.31KK341 pKa = 11.67LNIKK345 pKa = 9.98YY346 pKa = 10.27EE347 pKa = 4.17YY348 pKa = 10.2ISWDD352 pKa = 3.64EE353 pKa = 4.03IEE355 pKa = 4.14
MM1 pKa = 7.62KK2 pKa = 10.41NNNILKK8 pKa = 9.99IFLCVMVSLVVMCSGCVSSSSSDD31 pKa = 3.41SADD34 pKa = 3.05DD35 pKa = 3.64VVVEE39 pKa = 4.45KK40 pKa = 10.42TSTNDD45 pKa = 2.94NTEE48 pKa = 3.81PKK50 pKa = 10.21EE51 pKa = 4.06NEE53 pKa = 3.79EE54 pKa = 4.35TKK56 pKa = 10.86DD57 pKa = 3.48DD58 pKa = 4.16EE59 pKa = 4.51KK60 pKa = 11.22PKK62 pKa = 10.07TEE64 pKa = 4.02EE65 pKa = 3.9PVEE68 pKa = 4.13EE69 pKa = 4.53GLPDD73 pKa = 3.32EE74 pKa = 4.55YY75 pKa = 11.56VAVFYY80 pKa = 10.15EE81 pKa = 4.59AYY83 pKa = 8.33STKK86 pKa = 10.64SFDD89 pKa = 3.16GSYY92 pKa = 10.82GKK94 pKa = 10.65EE95 pKa = 3.43EE96 pKa = 3.88AADD99 pKa = 3.68GKK101 pKa = 10.92KK102 pKa = 10.63YY103 pKa = 10.71LIVDD107 pKa = 3.56ITVYY111 pKa = 11.17NNGYY115 pKa = 10.52DD116 pKa = 3.9EE117 pKa = 5.33ISVNPLYY124 pKa = 10.75FDD126 pKa = 5.56AIVDD130 pKa = 4.01GVEE133 pKa = 3.57YY134 pKa = 10.2DD135 pKa = 3.8YY136 pKa = 11.6SIDD139 pKa = 4.09SYY141 pKa = 11.77SLEE144 pKa = 4.03NDD146 pKa = 3.16LDD148 pKa = 4.26SVNLRR153 pKa = 11.84NGGKK157 pKa = 8.49TSGSLAFEE165 pKa = 4.28IPEE168 pKa = 4.84DD169 pKa = 3.78SSSTYY174 pKa = 10.44SIEE177 pKa = 4.1YY178 pKa = 10.58NPITWDD184 pKa = 3.43KK185 pKa = 11.53LNIIYY190 pKa = 10.59NPEE193 pKa = 3.93DD194 pKa = 3.51VDD196 pKa = 4.04IEE198 pKa = 4.25EE199 pKa = 4.37AQKK202 pKa = 10.99AAASSSNTGAEE213 pKa = 3.93KK214 pKa = 9.82STPTTSNDD222 pKa = 3.34DD223 pKa = 3.62TDD225 pKa = 4.07SDD227 pKa = 4.3SVNIITAATVTEE239 pKa = 4.19SFNYY243 pKa = 10.53NIGSYY248 pKa = 9.91EE249 pKa = 3.96YY250 pKa = 10.27TRR252 pKa = 11.84EE253 pKa = 4.08APSGKK258 pKa = 8.63TYY260 pKa = 10.76LITYY264 pKa = 10.17ISIEE268 pKa = 3.94NNGYY272 pKa = 10.5DD273 pKa = 4.35EE274 pKa = 5.39ISTSPLYY281 pKa = 10.63FDD283 pKa = 6.1AIVDD287 pKa = 4.11GVGYY291 pKa = 10.33SYY293 pKa = 11.73ASDD296 pKa = 3.52TYY298 pKa = 11.27SLEE301 pKa = 3.93NHH303 pKa = 6.55LEE305 pKa = 4.15SVDD308 pKa = 3.6LRR310 pKa = 11.84NGGKK314 pKa = 8.47TSGLLIYY321 pKa = 7.7EE322 pKa = 4.23IPKK325 pKa = 10.81GSDD328 pKa = 3.18SYY330 pKa = 11.26EE331 pKa = 4.09LEE333 pKa = 4.15YY334 pKa = 11.3DD335 pKa = 4.17PISWDD340 pKa = 3.31KK341 pKa = 11.67LNIKK345 pKa = 9.98YY346 pKa = 10.27EE347 pKa = 4.17YY348 pKa = 10.2ISWDD352 pKa = 3.64EE353 pKa = 4.03IEE355 pKa = 4.14
Molecular weight: 39.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7DQW1|D7DQW1_METV3 Uncharacterized protein OS=Methanococcus voltae (strain ATCC BAA-1334 / A3) OX=456320 GN=Mvol_1583 PE=4 SV=1
MM1 pKa = 7.82ASNKK5 pKa = 9.66PLGKK9 pKa = 10.15KK10 pKa = 9.18IRR12 pKa = 11.84LAKK15 pKa = 10.44ALNQNRR21 pKa = 11.84RR22 pKa = 11.84VPLFAVAKK30 pKa = 8.49TKK32 pKa = 10.68GAVRR36 pKa = 11.84SHH38 pKa = 6.01PKK40 pKa = 7.76MRR42 pKa = 11.84NWRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 5.59TLKK50 pKa = 10.45KK51 pKa = 10.72
MM1 pKa = 7.82ASNKK5 pKa = 9.66PLGKK9 pKa = 10.15KK10 pKa = 9.18IRR12 pKa = 11.84LAKK15 pKa = 10.44ALNQNRR21 pKa = 11.84RR22 pKa = 11.84VPLFAVAKK30 pKa = 8.49TKK32 pKa = 10.68GAVRR36 pKa = 11.84SHH38 pKa = 6.01PKK40 pKa = 7.76MRR42 pKa = 11.84NWRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 5.59TLKK50 pKa = 10.45KK51 pKa = 10.72
Molecular weight: 5.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
496543 |
33 |
2506 |
299.5 |
33.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.34 ± 0.071 | 1.348 ± 0.035 |
5.755 ± 0.042 | 7.718 ± 0.07 |
3.714 ± 0.039 | 6.206 ± 0.058 |
1.443 ± 0.021 | 9.561 ± 0.056 |
9.278 ± 0.092 | 9.016 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.556 ± 0.03 | 7.398 ± 0.106 |
3.141 ± 0.034 | 2.06 ± 0.024 |
2.922 ± 0.045 | 6.137 ± 0.057 |
5.159 ± 0.063 | 6.342 ± 0.055 |
0.588 ± 0.015 | 4.318 ± 0.054 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
